Chapter: Genetics and Molecular Biology: Transcription,Termination, and RNA Processing
Transcription Termination at Specific Sites
Transcription Termination at Specific Sites
If transcription of different genes is to be
regulated differently, then the genes must be transcriptionally separated. Such
transcriptional isola-tion could be achieved without explicit transcriptional
barriers between
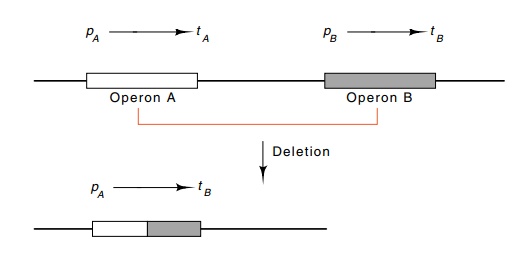
Figure
5.4 Fusion between two operons to
place some of the genes of operonB under control of the promoter of operon A.
The fusion removes the transcrip-tion termination signal tA and the promoter pB.
Transcription termination signals can be shown to
exist by several types of experiments. One that was first done in bacteria is
genetic. As mentioned earlier, transcriptional units are called operons. Even
though genes in two different operons may be located close to one another on
the chromosome, only by deleting the transcription termi-nation signal at the
end of one operon can genes of the second operon be expressed under control of
the first promoter (Fig. 5.4).
A second type of demonstration uses in vitro transcription. Radioac-tive RNA
is synthesized in vitro from a
well-characterized DNA template and separated according to size on
polyacrylamide gels. Some templates yield a discrete class of RNA transcripts
produced by initiation at a promoter and termination at a site before the end
of the DNA molecule. Thus these templates must contain a transcription
termination site. Of course, cleavage of an RNA molecule could be mistaken for
termination.
Related Topics