Chapter: Genetics and Molecular Biology: Transcription,Termination, and RNA Processing
S1 Mapping to Locate 5’ and 3’ Ends of Transcripts
S1 Mapping to Locate 5’ and 3’ Ends of
Transcripts
Early in the investigation of a gene and its
biological action, it is helpful to learn its transcription start and stop
points. One simple method for doing this is S1 mapping, which was developed by
Berk and Sharp. S1 is the name of a nuclease that digests single-stranded RNA
and DNA. S1 mapping shows on the DNA the endpoints of homology of RNA
mole-cules. That is, the method can map the 5’ and 3’ ends of the RNA. Consider
locating the 5’ end of a species of RNA present in cells. Messenger RNA is
isolated from the organism, freed of contaminating protein and DNA, then
hybridized to end-labeled, single-stranded DNA that covers the region of the 5’
end. After hybridization, the remaining single-stranded RNA and DNA tails are
removed by digestion with S1 nuclease. The exact size of the DNA that has been
protected from nuclease digestion can then be determined by electrophoresis on
a DNA sequencing gel (Fig. 5.10). This size gives the distance from the labeled
end of the DNA molecule to the point corresponding to the 5’ end of the RNA
molecule.
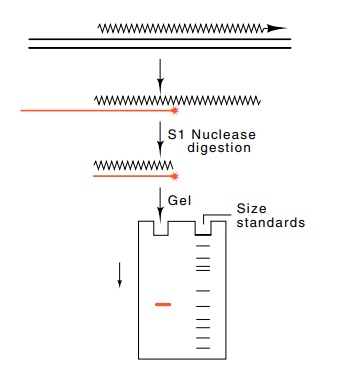
Figure
5.10 S1 mapping to locatethe 5’
transcription start site. RNA extracted from cells is hybridized to a
radioactive fragment covering the transcription start region, then digested
with S1 nuclease. The de-natured complex is subjected to electrophoresis
alongside size standards.
Related Topics