Chapter: Genetics and Molecular Biology: Transcription,Termination, and RNA Processing
A Common Mechanism for Splicing Reactions
A Common Mechanism for Splicing Reactions
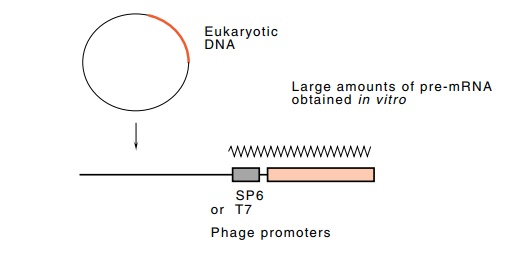
One early difficulty in studying splicing of mRNA
was obtaining the RNA itself. Cells contain large amounts of rRNA, but most of
the mRNA has been processed by splicing. Further, only a small fraction of the
unspliced pre–mRNA present at any moment is from any one gene. One convenient
source of pre-mRNA for use in splicing reactions came from genetic engineering.
The DNA for a segment of a gene containing an intervening sequence could be
placed on a small circular plasmid DNA molecule that could be grown in the
bacterium Escherichia coli and easily
purified. These circles could be cut at a unique location and then they could
be transcribed in vitro from special
phage promoters placed just upstream of the eukaryotic DNA (Fig. 5.21). By this
route, large quantities of unspliced substrate RNA could be obtained.
Figure
5.21 The use of SP6 or T7 phage
promoters on small DNA moleculesto generate sizeable amounts in vitro of RNAs suitable for study of
splicing reactions.
Figure
5.22 The two classes of self–splicing
RNA and the pathway of nuclearmRNA splicing, all drawn to emphasize their
similarities.
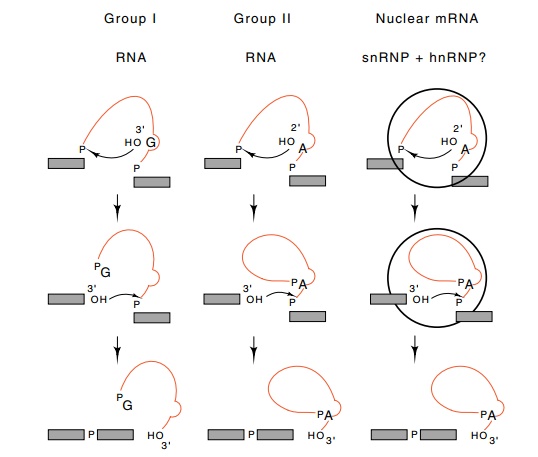
Both of the two self-splicing reactions and the
snRNP catalyzed splicing reactions can be drawn similarly (Fig. 5.22). In the
self-splicing cases a hydroxyl from a guanosine nucleotide or an adenine in the
chain attacks the phosphodiester and a transesterification ensues in which the
5’ end of the RNA is released. For the Group I self-splicing reaction, a tail
is formed, and for the Group II and mRNA splicing reactions, a ring with a tail
is formed. Then a hydroxyl from the end of the 5’ end of the molecule attacks
at the end of the intervening sequence and another transesterification reaction
joins the head and tail exons and releases the intron.
In the case of pre–mRNA splicing, the same reaction
occurs, but it must be assisted by the snRNP particles. In some intervening
sequences of yeast, internal regions are involved in excision and bear some
resem-blance to portions of the U1 sequences.
The similarities among the splicing reactions
suggest that RNA was the original molecule of life since it can carry out the
necessary functions on its own, and only later did DNA and protein evolve. The
splicing reactions that now require snRNPs must once have proceeded on their
own.
Related Topics