Chapter: Genetics and Molecular Biology: Transcription,Termination, and RNA Processing
Splicing Reactions and Complexes
Splicing Reactions and Complexes
In addition to requiring U1 snRNP particles,
pre-mRNA splicing re-quires at least three other snRNPs, U2, U5, and U4/U6, as
well as a number of soluble proteins. Together these form a large complex that
can be observed in the electron microscope, and which can be biochemi-cally
purified. The complex forms in the nucleus even while the RNA is being
elongated, and exons near the 5’ end of the RNA can be removed even before
synthesis of the RNA is complete. Formation of the complex requires that the
regions to which both U1 and U2 bind be present. Scanning by the splicing
apparatus from 5’ to the 3’ end may help explain the paradoxically high degree
of specificity to splicing. The donor and acceptor splice sites contain only
two essential nucleotides, too few to ensure specificity in RNA of random
sequence. It is likely that the spacing between introns and exons also helps
the splicing apparatus choose sites appropriately. One purification method of
spliceosomes is to synthesize substrate RNA in
vitro. This RNA is synthesized with ordinary nucleotides plus
biotin-substituted uridine. After the RNA has been added to a splicing extract,
the biotin can be used to fish out this RNA selectively with streptavidin bound
to a chromatography column. Along with the RNA are found the U1, U2, U4/U6, and
U5 snRNAs.
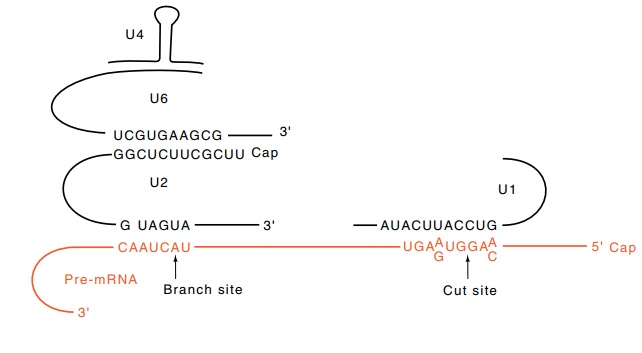
The reaction of the mammalian splicing components
is at least partly ordered. U1 binds by base pairing to the 5’ splice site and
U2 binds by base pairing to a sequence within the intervening sequence
containing a nucleotide called the branch point that participates in the
splicing reaction. The RNAs of U4 and U6 are extensively base paired whereas a
shorter region of base pairing is formed between the U6 and U2 RNAs (Fig.
5.19). In the course of the splicing reaction the U4 particle is released
first. Splicing in yeast is similar to that found in mammals, but
Figure
5.19 Base pairing among U1, U2, U4,
U6, and the pre-mRNA showingthe branch site and the 5’ cut site.
differs in many small details. The same snRNPs are
involved, but most of the U RNAs are considerably larger than their mammalian
counter-parts. Only U4 and U6 are closely homologous in both organisms.
Sometimes extensive splicing is required to
regenerate a single intact mammalian gene. For example, there are genes
containing one million bases and 60 splice sites. Only a few genes in yeast are
spliced, and they contain only a few introns. Great specificity is required in
order that all of the splicing reactions proceed with sufficient fidelity that
most of the pre-messenger RNA molecules ultimately yield correctly spliced
mes-senger RNA. In part, we still do not know the reasons for the high
fidelity. Although a consensus sequence at the 5’ and 3’ splice sites can be
derived by aligning many splice sites, there are only two invariant and
essential nucleotides present in each of the sites. This hardly seems like
enough information to specify correct splicing.
Once in vitro
splicing reactions could be performed with unique substrates, it was
straightforward to examine the products from the reactions. Amazingly, the
sizes of the products as determined by electrophoretic separations did not add
up to the size of the substrate pre-mRNA. The structures were then determined
by chemical means and by electron microscopy of the resultant RNA molecules.
The excised RNA was found to be in a lariat form.
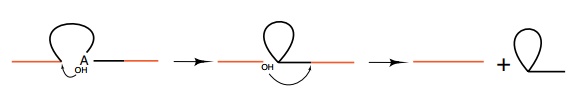
This results from the reaction of the nucleotide at
the branch point within the intron attacking the phosphodiester at the 5’
splice site. Subsequent attack of the 3’-OH at the 5’ splice site on the
phosphodiester bond at the 3’ splice site releases the intron in a lariat form
and completes the splicing process. The freed introns in lariat form are
rapidly degraded within the nucleus.
Related Topics