Chapter: Genetics and Molecular Biology: Transcription,Termination, and RNA Processing
Involvement of the U1 snRNP Particle in Splicing
Involvement of the U1 snRNP Particle in Splicing
Initially, progress in the study of the biochemical
mechanisms of splicing was slow. One of the first clues about the mechanism of
splicing came from the sequencing of many splice sites. It was noticed that the
sequence flanking the 5’ splice site was closely complementary to the sequence
of RNA found at the 5’ end of an RNA found in the U1 class of small nuclear
ribonucleoprotein particles called snRNPs. In addition to the U1 particles, U2,
U4, U6, and others exist, each containing 90 to 150 nucleotides and about 10
different proteins. The complementarity be-tween U1 RNA and the splice site of
pre-mRNA suggested that base pairing occurred between the two during splicing.
Stronger evidence for the proposal was the discovery by Steitz and Flint that
splicing in nuclei could be blocked by antibodies against U1 particles.
The experiment showing inactivation of splicing by
anti-U1 antibod-ies is not definitive since their specificity may not be high
and because other antibodies could also be present. One ingenious method for
specifically inactivating U1 particles was to remove nucleotides from their 5’
ends with RNAse H. This enzyme digests RNA from RNA-DNA
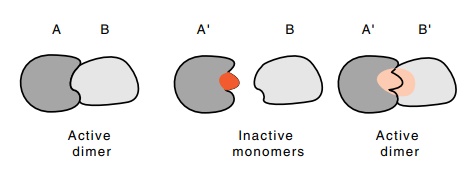
Figure
5.16 How compensating mutations in two
interacting structures canrestore activity.
duplexes. DNA oligonucleotides complementary to the
5’ end of U1 RNA were hybridized under gentle conditions to U1 particles in
cell extracts and then RNAseH was added. These treatments left the extracts
unable to catalyze intron removal whereas extracts which had received
oligonu-cleotides of different sequence were not inhibited.
One of the nicest demonstrations of biologically
significant interac-tions between two macromolecules are those in which
compensating mutations can be utilized. First, a mutation is isolated that
interferes with a particular interaction between A and B (Fig. 5.16). That is,
A’ no longer interacts with B in vivo.
Then a compensating mutation is isolated in B that potentiates the in vivo interaction between A’ and B’.
The isolation of the mutation and the compensating mutations in components of
the splicing apparatus could not be done using tradi-tional genetics tools
since the splicing reactions occur in animals for which only rudimentary
genetics exists. Genetic engineering methods had to be utilized.
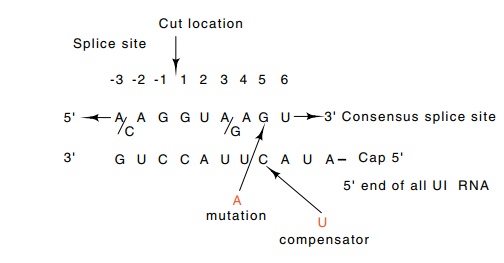
Figure 5.17 The consensus of the sequence at the 5’end of an exon-intronjunction. The sequence is highly complementary to the 5’ end of U1 RNA. Alterations in the splice sequence reduce splicing, and compensating alterations as shown in the U1 sequence restore splicing.
Two steps are necessary to perform the experiment.
The first is inducing the cells to synthesize messenger with an altered splice
site as well as synthesize U1 RNA with an altered sequence, and the second is
assaying for splicing of the special messenger in the presence of the normal
cellular levels of messenger and pre-messenger RNAs. Weiner introduced into the
cells a segment of adenovirus sequence coding for the E1a protein with
wild-type or variant 5’ splice sites as well as the gene for an altered U1.
Only when DNA was introduced that encoded a variant U1 gene that compensated
for the splice site mutation and restored Watson-Crick base pairing across the
region was the variant splice site utilized (Fig. 5.17).
The use of an adenovirus sequence for the experiment permitted altering the splicing of a messenger which was not important to the cells. Similarly, introducing a new gene for U1 RNA avoided disruption of the ongoing cellular splicing processes. Finally, examining splicing in the E1a gene from the virus increased the sensitivity since this gene contains three different 5’ splice sites, each utilizing the same 3’ splice site. Damaging the activity of one 5’ splice site diverted splicing to the other splice sites, whereas had there been only one 5’ splice site, altering it might only have slowed kinetics of the splicing process without chang-ing the actual amounts of RNA eventually spliced.
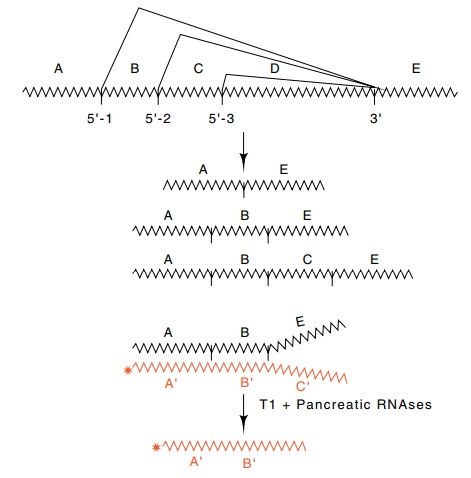
Figure 5.18 The pattern of splicing in adenovirus. The 5’splice sites separatingregions A, B, and C are all joined to the 3’ splice site separating regions D and E. Thus the three RNAs shown can be produced. The amount of the species ABE can be determined by hybridizing to radioactive RNA complementary to ABC and digesting with T1 and pancreatic RNAses. The amount of product of length AB is determined by electrophoresis.
An RNAse protection assay was used to monitor the
splicing. RNA was isolated from the cells and hybridized with radioactive RNA
com-plementary to the adenovirus mRNA. The regions of the RNA which are not
base paired are sensitive to T1 and pancreatic RNAses and are digested away
(Fig. 5.18). This leaves a radioactive RNA molecule whose size indicates the
splice site utilized. A variant sequence at the middle E1a splice site
eliminated splicing from this location, but the introduc-tion of the variant U1
gene to the cells containing the compensating mutation restored use of this
splice site.
Related Topics