Chapter: Genetics and Molecular Biology: RNA Polymerase and RNA Initiation
Melted DNA Under RNA Polymerase
Melted DNA Under RNA Polymerase
One of the first steps of transcription is the
binding of RNA polymerase to the proper sequence on the DNA. The best evidence
at present supports the notion that RNA polymerase reads the sequence of the
DNA and identifies the promoter in a double-stranded unmelted structure. It is
theoretically possible that the bases of the growing RNA could be specified by
double-stranded unmelted DNA, but it seems vastly easier for these bases to be
specified by Watson-Crick base pairing to a partially melted DNA duplex.
Direct experimental evidence shows that during the
initiation proc-ess, RNA polymerase melts at least 11 base pairs of DNA. For
example, positions on adenine rings normally occupied in base pairs become
available for chemical reaction if the pairs are disrupted, and their exact
positions along a DNA molecule can then be determined by methods analogous to
those used in DNA sequencing. Results obtained from this type of measurement
reveal that 11 base pairs of DNA from about the middle of the Pribnow box to
the start site of transcription are melted when the RNA polymerase binds to a
promoter.
A different method has also been used to measure
the amount of DNA that is melted by the binding of RNA polymerase. This method
consists of binding RNA polymerase to a nicked circular DNA molecule, sealing
the nick with polymerase still bound, and determining the change in the
supercoiling generated by the presence of the polymerase. If we assume that the
melted DNA strands are held parallel to the helix axis, this method yields 17 base
pairs melted. The problem is that no method has been developed to determine
whether the melted region contains any twist. If it does, then the size of the
region that is melted cannot be precisely determined (Fig. 4.23).
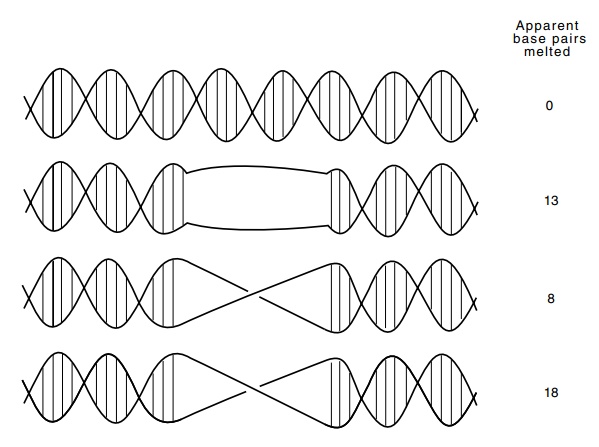
Figure
4.23 Topological measurements cannot
give the exact number of basepairs opened by binding of a protein. In each of
the three cases, 13 base pairs are broken, but only if the melted region is not
twisted does the DNA contain one less twist.
The melting of 10 to 15 base pairs of DNA under physiological conditions requires appreciable energy because this length of oligomers hybridize together with a very high binding
constant. The tight hybridi-zation derives from the same source that pushes DNA
to the double helical structure, base stacking interactions and hydrogen bonds.
Since a large amount of energy is required to melt the DNA and a limited amount
of energy is available from the binding of RNA polymerase, thermal motion in
the solution provides the activation energy for the melting. Once this occurs,
polymerase binds tightly to parts of the separated strands and maintains the
bubble. At low temperatures, and therefore lower thermal motion, an RNA
polymerase-DNA duplex is much less likely to possess the requisite activation
energy, and the melting rate at lower temperatures is much reduced. At 0°
virtually no RNA polymerase bound to phage T7 DNA is able to initiate in
reasonable periods of time, while at 30° almost 100% has isomerized and is able
to initiate within a few minutes. Similarly, the melting rate is affected by
the salt concentration.
Related Topics