Chapter: Genetics and Molecular Biology: RNA Polymerase and RNA Initiation
Enhancer-Binding Proteins
Enhancer-Binding Proteins
Some enhancer-binding proteins like the
glucocorticoid receptor protein have been purified and studied in vitro. Other enhancer-binding
proteins, like the GAL4 and GCN4 proteins from yeast, can be engineered and
studied in vivo without ever
purifying the protein. Both types of studies indicate that enhancer proteins
possess multiple independent domains. The DNA-binding domain of the GAL4
enhancer protein can be replaced with the DNA-binding domain of a bacterial
repressor protein, LexA (Fig. 4.14). When the LexA-binding sequence is placed
in front of some other yeast gene, the gene acquires the ability to be induced
by the GAL4-LexA hybrid protein when galactose is present. The glucocorticoid
receptor protein possesses DNA-binding, steroid-binding, and activation
domains. These too can be separated and interchanged.
The regions of enhancer proteins necessary for
activation can be explored. By progressively deleting protein from an
activating domain, both Ptashne and Struhl have found that stretches of
negatively charged amino acids on some activator proteins are required for
activation. In
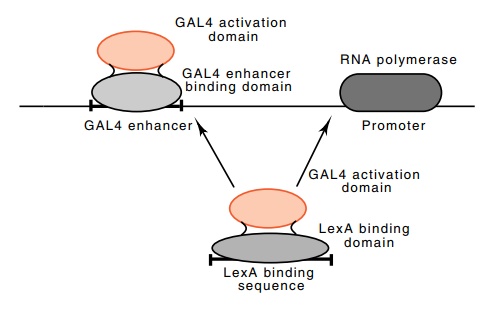
Figure
4.14 Hybrid enhancer proteins can
activate transcription if they areheld in the right areas of DNA.
GCN4, two such regions are required for full
activation abilities. It appears necessary that these negatively charged amino
acids all lie on one face of an alpha helix. The other side of the helix can be
hydropho-bic. Activating helices can be designed de novo if they follow these principles, but if the charged amino
acids are shuffled, they do not activate. Other structures in addition to
negatively charged surfaces of α-helices
also function to activate RNA polymerase. Some enhancer-binding proteins lack
significant negatively charged regions and instead possess large quantities of
proline or glutamine.
Many enhancer proteins may be relatively simple.
They can possess nearly independent domains for DNA-binding, for binding a
small molecule like a hormone, and for activating RNA polymerase or the basal
machinery. The binding of a hormone may unmask the DNA-bind-ing domain or the
activation domain of the protein. In some cases the activating domain may be
little more than a high concentration of negative charge whose interaction with
TFIID activates transcription.
Activation in eukaryotes has to contend with the
presence of histones tightly-bound to the DNA. Undoubtedly, their presence
interferes with transcription. Some activator proteins therefore overcome the
repres-sive effects of bound histones. Other activator proteins can be expected
to go beyond overcoming repression, and will stimulate transcription.
The number of different enhancer-binding proteins
appears to be remarkably small in nature. Repeatedly, researchers are finding
that some of the enhancer proteins from one gene are highly similar to one that
controls another gene, either in that same organism or in a different organism.
Not only are the sequences of such proteins similar, but they are functionally
interchangeable. Heterologous in vitro
transcription systems can be constructed in which enhancer proteins from yeast
activate transcription from a human system. The yeast GCN4 protein is similar
to the AP-1, c-myc, c-jun, and c-fos proteins. AP-1 is a mammal
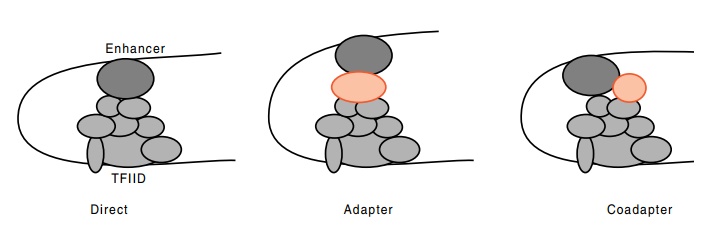
Figure
4.15 Enhancer proteins can interact
with the TFIID complex directlyor through other proteins, shown in red.
Many of the enhancer-binding proteins interact with
the TFIID complex. Because gene regulation is so important to the cell and
because many different genes in a cell must be regulated, we can expect a wide
diversity of interaction modes (Fig. 4.15). The enhancer-binding proteins can
interact directly, via adapters, or in cooperation with coadapters.
Additionally, proteins that are part of the TFIID complex or the other proteins
may be required for some interactions and interfere with others. That is, a
protein may play an activating role in the expression of some genes and a
repressing role in the expression of others.
Related Topics