Chapter: Genetics and Molecular Biology: RNA Polymerase and RNA Initiation
Multiple but Related Subunits in Polymerases
Multiple but Related Subunits in Polymerases
How can one be sure that an enzyme contains
multiple subunits? One of the best methods for detection of multiple species of
polypeptides in a sample is electrophoresis through a polyacrylamide gel. If
the protein has been denatured by boiling in the presence of the detergent
sodium dodecyl sulfate, SDS, and the electrophoresis is performed in the
pres-ence of SDS, polypeptides separate according to size. This results from
the fact that the charged SDS anions that bind to the polypeptides completely
dominate the charge as well as force all polypeptides to adopt a rodlike shape
whose length is proportional to the molecular
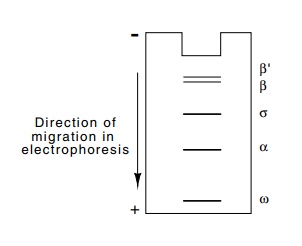
Figure
4.8 The polypeptide bandpattern found
by SDS polyacry-lamide gel electrophoresis of purified E. coli RNA polymerase
weight of
the protein. Therefore, in most cases two polypeptides of the same size will
migrate at the same rate and two of different molecular weight will migrate at
different rates. Following electrophoresis, the positions of proteins in the
gel can be visualized by staining. Each band on a gel derives from a
different-sized polypeptide species.
When
purified E. coli RNA polymerase is subjected to SDS polyacry-lamide gel
electrophoresis, five distinct bands are seen (Fig. 4.8). The mere presence of
multiple polypeptides in purified enzyme doesn’t prove that all the peptides
are necessary for activity. Do all the bands on the gel represent subunits of
RNA polymerase or are some of the bands extraneous proteins that adventitiously
copurify with RNA po-lymerase? A reconstitution experiment provides the most
straightfor-ward demonstration that the four largest polypeptides found in RNA
polymerase are all essential subunits of the enzyme. The four bands from an SDS
polyacrylamide gel are cut out, the proteins eluted, and SDS removed. RNA
polymerase activity can be regained only if all four of the proteins are
included in the reconstitution mixture.
RNA
polymerase from E. coli consists of subunits β’ and β of molecu-lar weights 155,000
and 151,000, two subunits of α whose
molecular weight is 36,000, a low molecular weight subunit ω whose presence is not necessary
for activity, and one somewhat less tightly-bound subunit, σ of 70,000 molecular weight.
Measurement of the amounts of each ofthe five proteins on SDS polyacrylamide
gels shows that the enzyme contains two copies of the α subunit for every single copy of
the others, that is, the subunit structure of RNA polymerase is σα2ββ’ω.
The reconstitution experiments permit pinpointing
the actual target of rifamycin. RNA polymerase from rifamycin-sensitive and
rifamycin-resistant cells is subjected to SDS polyacrylamide gel
electrophoresis. Then reconstitution experiments with the two sets of proteins
can be performed in all possible combinations to determine which of the four
subunits from the rifamycin-resistant polymerase confers resistance to the
reconstituted enzyme. The β subunit
was found to be the target of rifamycin.
If we view the RNA polymerase as a biochemical
engine, then it is reasonable to expect each subunit to have a different
function. As discussed below, rifamycin inhibits initiation by RNA polymerase,
but
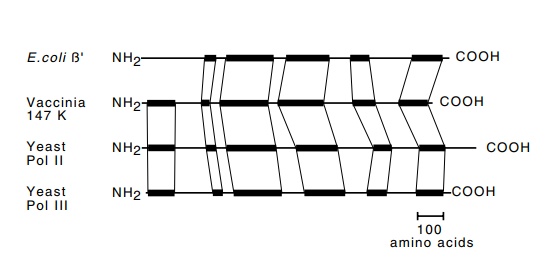
Figure
4.9 The dark bands indicate the
similarities among theβ’subunits
ofRNA polymerase from E. coli, vaccinia virus, and yeast
polymerases II, and III.
A different antibiotic, streptolydigin, has also been
found to inhibit RNA polymerase. This blocks elongation steps, and therefore we
might have expected to find a subunit other than β to be the target of this drug. Alas, however, the β subunit is also the target of streptolydigin. Some
spe-cialization exists. The β subunit
binds ribonucleotides and possesses the catalytic site while the β’ subunit binds DNA. Most likely the larger two
subunits are comprised of a number of domains, each playing a different role in
the initiation and elongation of RNA. Evolution seems to have conserved the
structures and functions of some of these different do-mains. The larger
subunits from prokaryotic and the three types of eukaryotic RNA polymerase all share
significant homology. Regions of homology are also found amongst the other
subunits as well.
The combined molecular weights of the subunits of
RNA polymerase total nearly one half million, but from a mechanistic viewpoint
it is not at all clear why the polymerase should be so large. Phage T7, which
grows in E. coli, encodes its own RNA polymerase, and this enzyme has a
molecular weight of only about 100,000. Apparently the actual RNA initiation
and elongation steps do not require an enzyme as large as the E.
coli polymerase. Perhaps the large size of the cellular polymerasespermits
them to initiate from a wider variety of promoters and to interact with a
variety of auxiliary regulatory proteins.
The eukaryotic RNA polymerases are also large and
possess multiple subunits. RNA polymerase II from many different organisms has
been shown to contain 12 different polypeptides. The largest three are
ho-mologous to β’, β, and α of the E. coli RNA polymerase. Fig. 4.9 shows the shared homology of the β’ subunit among the E. coli, vaccinia virus, and Saccharomyces
cerevisiae polymerases II and III. The RNA polym-erases I and III possess
five subunits in common with RNA polymerase II.
The eukaryotic polymerases contain more subunits
than the E. coli RNA polymerases. Part
of the differences may merely be in the tightness with which subunits cling
together. For example, a protein from E.
Coli that is involved with RNA chain
termination, the nusA gene product,
could be considered a part of RNA polymerase because it binds to the polymerase
after initiation has occurred and after the σ subunit has been released from the core complex of β, β’, and α. Since however, it does not copurify with the RNA
chain elongating activity that is con-tained in the core, it usually is not classified
as part of RNA polymerase. Some of the peptides in the eukaryotic polymerase
might just stick together more tightly.
One notable difference between the prokaryotic and
eukaryotic po-lymerases is that the largest subunit of RNA polymerase II possesses
at its C-terminal end a heptad of (Tyr-Ser-Pro-Thr-Ser-Pro-Ser) repeated 25 to
50 times. This C-terminal domain, or CTD, can be multiply phosphorylated by any
of several proteins known to activate transcrip-tion on various promoters. It
appears that the unphosphorylated form of the CTD helps RNA polymerase bind and
interact with the auxiliary

proteins necessary for transcription initiation,
but that the CTD must be phosphorylated before it can release from the
initiation proteins and allow the polymerase to elongate freely. Cells
constructed so as to lack the CTD, or cells in which the CTD is too short are
sick or inviable.
Related Topics