Chapter: Genetics and Molecular Biology: RNA Polymerase and RNA Initiation
The RNA Polymerase in Escherichia coli
The RNA Polymerase in Escherichia coli
Once biochemists could assay and purify an RNA
polymerase from E. coli, it was important to know the
biological role of the enzyme. For example, the bacterial cell might possess
three different kinds of RNA polymerase: one for the synthesis of messenger
RNA, one for the synthesis of tRNA, and one for the synthesis of ribosomal RNA.
If that were the case, much effort could have been wasted in studying in vitro transcription from a gene if
the wrong RNA polymerase had been used. Enzymologists’ failures to find more
than one type of RNA polymerase in E.
coli were no proof that more did not
exist, for, as we have seen, detection of an enzyme in cells can be difficult.
In other words, what can be done to determine the biological role of the enzyme
that can be detected and purified?
Fortunately, a way of determining the role of the E. coli
RNA polym-erase finally appeared. It was in the form of a very useful
antibiotic, rifamycin, which blocks bacterial cell growth by inhibiting
transcription initiation by RNA polymerase. If many cells are spread on agar
medium containing rifamycin, most do not grow. A few do, and these
rifamycin-resistant mutants grow into colonies. Such mutants exist in
populations of sensitive cells at a frequency of about 10-7.
Examination of the resistant mutants shows them to be of two classes. Mutants
of the first class are resistant because their cell membrane is less permeable
to rifamycin than the membrane in wild-type cells. These are of no interest to
us here. Mutants of the second class are resistant by virtue of an alteration
in the RNA polymerase. This can be demonstrated by the fact that the RNA
polymerase purified from such rifamycin-resistant cells has become resistant to
rifamycin.
Since rifamycin-resistant cells
now contain a rifamycin-resistant polymerase, it would seem that this
polymerase must be the only type present in cells. Such need not be the case,
however. Consider first the hypothetical possibility that cells contain two
types of RNA polymerase, one that is naturally sensitive to rifamycin, and one
that is naturally resistant. We might be purifying and studying the first
enzyme when we should be studying the naturally resistant polymerase. This
possibility of this situation can be excluded by showing that rifamycin
addition to cells stops all RNA synthesis. Therefore cells cannot contain a
polym-erase that is naturally resistant to rifamycin. A second possibility is
that cells contain two types of polymerase and both are sensitive to
rifamy-cin. Because mutants resistant to rifamycin can be isolated, both types
of polymerase would then have to be mutated to rifamycin resistance. Such an
event is exceedingly unlikely, however. The probability of mutating both
polymerases is the product of the probability of mutating either one. From
other studies we know that the mutation frequency for such an alteration in an
enzyme is on the other of 10-7. Therefore, the probability of
mutating two polymerases to rifamycin resistance would be about (10-7)2
(Fig. 4.6), which is far below the frequency of 10-7 that is
actually observed.
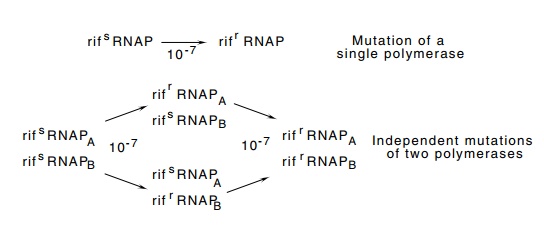
Figure
4.6 Mutation of a single RNA
polymerase to rifamycin resistanceoccurs at a frequency of 10-7, whereas
if two independent mutational events are required to mutate
two differet polymerases to rifamycin resistance, the frequency is 10-7× 10-7= 10-14.
Thus far,
then, we know these facts: The target of rifamycin is a single type of RNA
polymerase in bacteria. This RNA polymerase synthesizes at least one essential
class of RNA, and this polymerase is the one that biochemists purify. How do we know that this RNA polymerase
synthe-sizes all the RNAs? Careful physiological experiments show that
rifamy-cin addition stops synthesis of all classes of RNA, mRNA, tRNA, and
rRNA. Therefore the same RNA polymerase molecule must be used for the synthesis
of these three kinds of RNA, and this RNA polymerase must be the one that the
biochemists purify.
Unfortunately there is an imperfection in the
reasoning leading to the conclusion that E.
coli cells contain only a single type
of RNA polym-erase molecule. That imperfection came to light with the discovery
that the prokaryotic RNA polymerase is not a single polypeptide but in fact
contains four different polypeptide chains. Therefore, the rifamycin experiment
proves that the same polypeptide is used by whatever polymerases synthesize the
different classes of RNA. Much more ardu-ous biochemical reconstruction
experiments have been required to exclude the possibility that bacteria contain
more than one single basic core RNA polymerase.
Related Topics