Chapter: Genetics and Molecular Biology: RNA Polymerase and RNA Initiation
Structure of Promoters
The Structure of Promoters
Some
proteins in the cell are needed in high quantities and their messenger RNAs are
synthesized at a high rate from active promoters. Other proteins are required
in low levels and the promoters for their RNAs have low activity. Additionally,
the synthesis of many proteins must respond to changing and unpredictable conditions
inside or out-side the cell. For this last class, auxiliary proteins sense the
various conditions and appropriately modulate the activities of the promoter.
Such promoters must not possess significant activity without the assis-tance of
an auxiliary protein. Not surprisingly, then, the sequences of promoters show
wide variations. Yet behind it all, there could be elements of a basic
structure or structures contained in all promoters.
As the
first few bacterial promoters were sequenced, considerable variation was
observed between their sequences, and their similarities could not be
distinguished. When the number of sequenced promoters reached about six,
however, Pribnow noticed that all contained at least part of the sequence
TATAAT about six bases before the start of mes-sengers, that is, 5’XXXTATAATXXXXXAXXXX-3’, with the messenger often beginning with A. This sequence is often called the
Pribnow box. Most bacterial promoters also possess elements of a second region
of conserved sequence, TTGACA, which lies about 35 base pairs before the start
of transcription. Examination of the collection of E. coliσ70
promoters reveals not only the bases that tend to be conserved at the -35 and
-10 regions, but also three less well conserved bases near the transcription
start point (Fig. 4.10). The figure also shows that the spacing between the -35
and -10 elements is not rigidly retained, al-though the predominant spacing is
17 base pairs. To a first approximation, the more closely a promoter matches
the -10 and -35 sequences and possesses the correct spacing between these
elements, the more active the promoter. Those promoters that utilize regulatory
proteins to activate transcription deviate from the consensus sequences in
these regions. These activating proteins must help the polymerase through steps
of the initiation process that it can do by itself on “good” promoters. The
promoters that are activated by other σ factors possess other
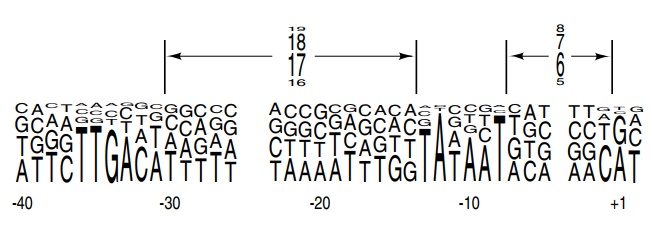
Figure
4.10 Consensus of allE. colipromoters in which the height of
a baseat each position is proportional to the frequency of occurrence of that
base. Similarly, the heights of the numbers represent the frequency of the
indicated spacings between the elements.
sequences in both the -35 and -10 regions. This
implies that the σ subunit contacts both regions of
the DNA. Direct experiments have shown this is the case. Perhaps the most
elegant was the generation of a hybrid promoter with a -35 region specific for
one type of σ and a -10 region specific for a
different σ. The hybrid promoter could be
activated only by a hybrid σ subunit
containing the appropriate regions from the two normal σ subunits.
Protection of promoters from DNAse digestion,
chemical modifica-tion, and electron microscopy have shown that polymerase
indeed is bound to the -35 and -10 regions of DNA before it begins transcription
and that it contacts the DNA in these areas. Experiments of the type described have
permitted direct determination of
Figure
4.11 The consensus sequences found inE.colipromoters
and thehelical structure of the corresponding DNA. The majority of the contacts
between RNA polymerase and DNA are on one side of the DNA.
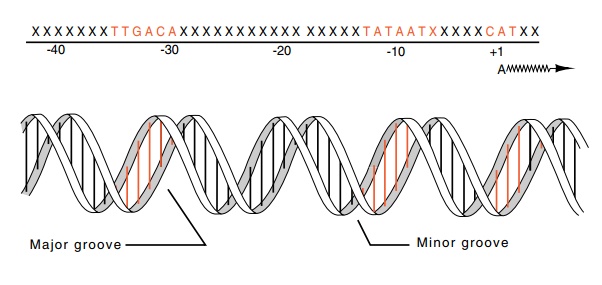
the bases and phosphates contacted in the promoter
region that E. coli RNA polymerase contacts. These areas are clustered in the regions
of the conserved bases in the -35, -10, and +1 areas (Fig. 4.11). These regions
all may be contacted by RNA polymerase binding to one face of the DNA. More
complicated experiments have attempted to answer the reverse question of which
polymerase subunits contact these bases.
Study of eukaryotic promoters reveals relatively
few conserved or consensus sequences. Most eukaryotic promoters possess a TATA
se-quence located about 30 base pairs before the transcription start site. In
yeast, however, this sequence is found up to 120 base pairs ahead of
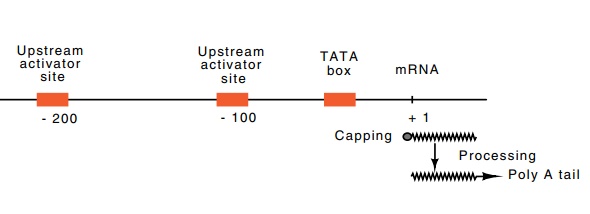
The eukaryotic minimal RNA polymerase II promoter
consists of the TATA box and additional nucleotides around the transcription
start
Figure
4.12 The binding order and approximate
location of the transcriptionfactors TFIIA, B, D, and E, and RNA pol II.
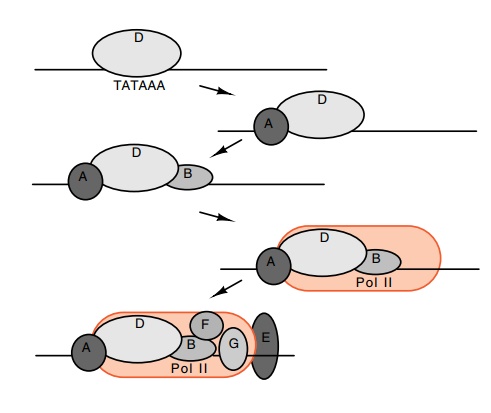
point. The apparatus required for initiation is the
polymerase plus at least six additional proteins or protein complexes (Fig.
4.12). The first to bind is TFIID. This is a complex of about ten different
proteins. The one that binds the TATA sequence is called TATA-binding protein,
and the others are called TATA associated factors or TATA associated proteins.
Although TFIID was first recognized as necessary for initiating transcription
by RNA polymerase II, at least the TATA-binding protein of TFIID is now known
to be required for initiation by all three types of RNA polymerase, I, II, and
III. At the promoters served by RNA polym-erase II, after TFIID binds, then
TFIIA and B bind followed by RNA polymerase II. After this TFIIE, F, G and
still others bind. The order of protein binding in vitro and the approximate location of binding of the proteins
was assayed using DNAse footprinting and the migration retardation assay. In
this assay, a piece of DNA about 200 base pairs long is incubated with various
proteins and then subjected to electro-phoresis under conditions that increase
the tightness of proteins bind-ing to the DNA. The DNA migrates at one rate,
and the protein-DNA complex migrates more slowly through the gel, thereby
permitting detection and quantitation of protein binding to DNA.
Related Topics