Chapter: Genetics and Molecular Biology: RNA Polymerase and RNA Initiation
DNA Looping in Regulating Promoter Activities
DNA Looping in Regulating Promoter Activities
DNA looping is a reasonable way enhancers can
interact with the transcription apparatus. The available data say that this,
indeed, is one of the ways they work. For example, an enhancer can be placed on
one DNA circle, and the promoter it stimulates on another DNA circle. When the
DNA rings are linked, the enhancer functions. This shows that the enhancer must
be close to the promoter in three-dimensional space. The linking experiment also
shows that a protein or signal doesn’t move down the DNA from the enhancer to
the promoter.
DNA looping solves two physical problems in gene
regulation. The first concerns space. Regulatory proteins must do two things.
They sense intracellular conditions, for example, the presence of a growth
hor-mone. They then must turn on or turn off the expression of only those genes
appropriate to the conditions. These responses require that a signal be
transmitted from a sensor part of the regulatory protein to the cellular
apparatus responsible for transcribing or initiating transcrip-tion from the
correct gene. The phrase “correct gene” is the key here. How can the regulatory
protein confine its activity to the correct gene?
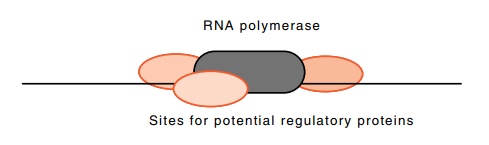
Figure
4.16 The limited number of sites
immediately adjacent to an RNApolymerase molecule.
The easiest, and virtually the only general way for
a regulatory protein to recognize the correct gene is for the protein to
recognize and bind to a DNA sequence near or within the correct gene.
If a regulatory protein is bound adjacent to an RNA
polymerase molecule or adjacent to an auxiliary protein required for initiating
transcription, we can imagine direct protein-protein contacts for
com-munication of the necessary signals. The space problem arises since only a
limited number of proteins may bind immediately adjacent to the transcription
initiation complex (Fig. 4.16). The limit seems to be two to four proteins.
Since the regulatory pattern of many genes is complex and likely to require the
combined influence of more than two or three regulatory proteins, we have a
problem.
How can more than a couple of proteins directly
influence the RNA polymerase? DNA looping is one answer. A regulatory protein
can be bound within several hundred or several thousand base pairs of the
initiation complex and directly touch the complex by looping the DNA. With DNA
looping a sizeable number of proteins can simultaneously affect transcription
initiation via multiple loops. Additional possibilities exist. For example,
proteins could regulate by helping or hindering loop formation or alternative
looping could exist in the regulation scheme of a gene.
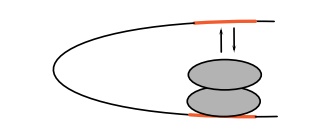
A second reason for DNA looping is the
cooperativity generated by a system that loops. Consider a system in which a
protein can bind to two DNA sites separated by several hundred base pairs and
then the proteins can bind each other, thus forming a DNA loop. A different
reaction pathway can also be followed. A molecule of the protein could bind to
one of the sites and a second molecule of the protein could bind to the first.
By virtue of the potential for looping, the concentration of the second protein
in the vicinity of the second DNA site has been increased.
Such a concentration change increases the occupancy
at the second site above the value it would have in the absence of looping.
Hence, the presence of one site and looping increases occupancy of the second
site. Such a cooperativity can substantially facilitate binding at low concen-trations
of regulatory proteins. It also eliminates any time lags upon gene induction
associated with diffusion of a protein to its DNA-binding site.
Increasing the local concentration of a regulatory protein near its binding site solves a serious problem for cells. Thousands of regulatory proteins must be present in a bacterial cell and
tens of thousands of regulatory proteins may have to be present in the nucleus
of some eukaryotic cells. Since the total protein concentration possible in the
cell or nucleus is limited to about 200 mg/ml, and since the same space must be
shared with the chromosome and housekeeping proteins as well, the concentration
possible for any one type of regulatory protein is strictly limited.
How then can the requisite binding of the
regulatory protein to its DNA target sequence be achieved? Basically, the
effective concentration of the protein must be high relative to the
dissociation constant from the site. If the affinity is too high, however, the
tightness of binding may make the dissociation rate from the site so slow that
the protein’s presence interferes with normal cellular activities like DNA
replication, recombination, and repair. A nice solution to these contradictory
re-quirements is to build a system in which the affinity of the protein for the
site is not too high, the overall concentration of the protein in the cell is
not too high, but the local concentration of the protein just in the vicinity
of the binding sites is high. DNA looping provides a simple mechanism for
increasing the local concentrations of regulatory pro-teins.
Related Topics