Chapter: The Diversity of Fishes: Biology, Evolution, and Ecology: Fish genetics
Fish genomics
Fish genomics
Genomics is the study of the entire DNA sequence of an organism, which in fishes includes the small mitochondrial genome and the enormous nuclear genome that can contain over a billion base pairs (bp). The earliest explorations of fish genomics were chromosome counts and karyotypes, but genomics now refers primarily to the intensive efforts to record the entire nuclear genome for many species. In the late 1990s, the technology applied to the Human Genome Project was redirected towards fishes and other vertebrates, and a cottage industry of fish genome projects is emerging. As of this writing, four fish genomes are completely described: Zebrafish (Danio rerio), Medaka (Oryzias sp.), and two pufferfishes. Carp (Cyprinus carpio), cichlid (genus Tilapia), and salmon (genus Oncorhynchus) are the subject of upcoming genome projects, and doubtless this list will be expanded over the next several decades.
Genome size
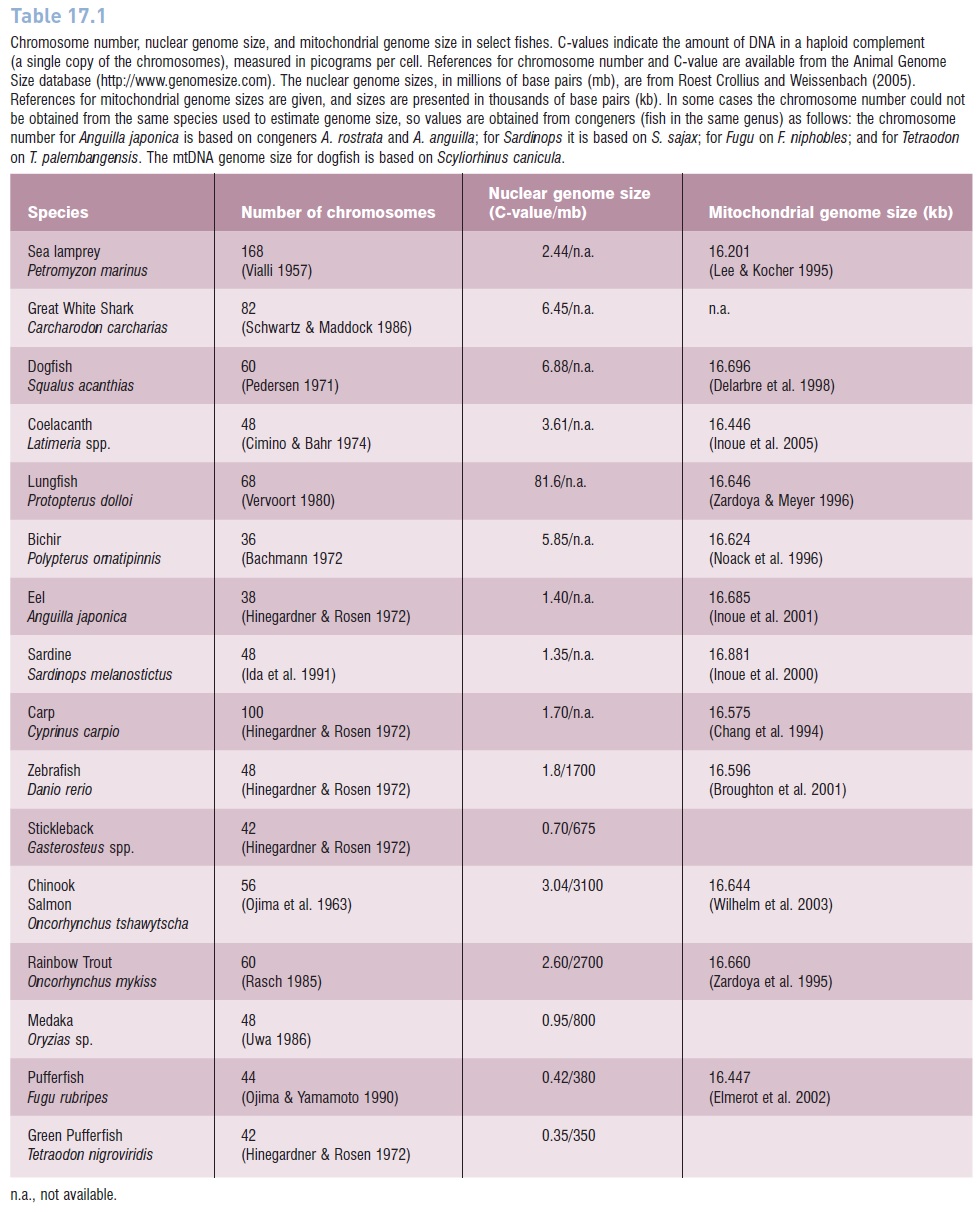
Fish genomes include 21 to 100+chromosomes in the nuclear genome (nDNA) with two copies of every gene in most (diploid) species. Some 58% of examined teleosts (334 out of 580 species) have 48 or 50 chromosomes (Naruse et al. 2004), and 48 is believed to be the ancestral state for ray-finned fishes. Despite this conservation of chromosome number, overall genome sizes can differ by more than two orders of magnitude, with the lungfish having the largest fish genome (81.6 pg of DNA in a set of chromosomes), the bichir having the largest actinopterygian (ray-finned fish) genome (5.85 pg), and the tetraodontiforms (such as pufferfish) having the smallest genomes (0.35 pg (Table 17.1), compared to 3.4 pg in the human genome. The number of chromosomes and genome size can vary even within a single genus (see the Oncorhynchusexamples in Table 17.1). Three trends in genome size are apparent:
1 There is a progressive reduction in DNA content from the earliest to the most advanced bony fishes.
2 Apart from lungfishes, elasmobranchs have the largest fish genomes, with 3–34 pg of DNA (Stingo & Rocco 2001). In contrast, the holocephalians (the other major group of cartilaginous fishes) have among the most compact genomes (1.2–1.9 pg; Venkatesh et al. 2005).
3 In general, freshwater fishes have larger genomes than marine fishes (Yi & Streelman 2005). This is attributed to smaller population sizes in freshwater fishes, which can reduce the power of natural selection to produce a compact genome.
It is notable that fishes with the most radically derived morphology (tetraodontiforms) have the smallest nuclear
genome. The pufferfish (genus Fugu) provided an ideal candidate for the first fish genome study, having much the same set of genes observed in mammals, but in a package eight times smaller than the human genome. The insights from the first round of fish genome studies are many, and highlights include:
❄Genomes are dynamic with many rearrangements between species and sometimes within species. Segments of the genome that are similar between fishes and mammals are rarely longer than four genes.
❄Previous estimates of the vertebrate genome ranged from 60,000 to 150,000 genes, but that number now appears to be 30,000–40,000 genes.
❄There are regions of the genome of unknown function that are high conserved (very similar) between fishes and mammals. High similarity between fishes and mammals indicates that these gene regions have important functions that are under strong natural selection.
Polyploidization and evolution
Polyploidization is the wholesale duplication of the nuclear genome, and most authorities agree that such an event lies near the base of the ray-finned fish (Actinopterygian) evolutionary tree. Such events are rare but important in the evolution of fishes. Ohno (1970) proposed that gene duplication is essential for major evolutionary innovations in vertebrates, as opposed to the single nucleotide mutations that can distinguish populations and species. In this view, the duplicated genes are under relaxed selection pressure, because there are now four copies (instead of two) available to get the job done. When polyploidy occurs, the original function of the gene can be maintained, freeing the extra copies to develop new functions (neofunctionalization), or they can double the capacity of a crucial metabolic pathway. Over tens of millions of years, some of the duplicated genes will prove to be redundant and lose their function in a process known as diploidization (returning to the diploid state). The few genes that are retained may allow evolutionary innovations.
Table 17.1
Chromosome number, nuclear genome size, and mitochondrial genome size in select fishes. C-values indicate the amount of DNA in a haploid complement (a single copy of the chromosomes), measured in picograms per cell. References for chromosome number and C-value are available from the Animal Genome Size database (http://www.genomesize.com). The nuclear genome sizes, in millions of base pairs (mb), are from Roest Crollius and Weissenbach (2005). References for mitochondrial genome sizes are given, and sizes are presented in thousands of base pairs (kb). In some cases the chromosome number could not be obtained from the same species used to estimate genome size, so values are obtained from congeners (fish in the same genus) as follows: the chromosome number for Anguilla japonica is based on congeners A. rostrata and A. anguilla; for Sardinops it is based on S. sajax; for Fugu on F. niphobles; and for Tetraodon on T. palembangensis. The mtDNA genome size for dogfish is based on Scyliorhinus canicula.
Dramatic support for Ohno’s model of evolution came from the Zebrafish (Danio rerio) genome, and the discovery of seven HOX genes (important regulators of morphological development during embryonic growth). In mammals there are four HOX genes on four chromosomes, whereas the Zebrafish lineage apparently had eight HOX genes on eight chromosomes, followed by the loss of one copy (Amores et al. 1998). The pufferfish genomes provide additional evidence of gene duplication and neofunctionalization.
When did this early genome duplication occur? The duplication does not appear in sturgeon or gar, but is shared by all surveyed teleosts (Hoegg et al. 2004). Hence this event must have occurred in the basal teleost lineage, on the order of 300–400 million years before present (mybp). This genome duplication may have provided a powerful toolkit for diversifi cation of the teleosts, by providing twice as many genes as existed in ancestral fishes (Roest Crollius & Weissenback 2005).
The ancient polyploidization in teleosts is not the only genome duplication in bony fishes. Additional wholegenome duplications have occurred in the ancestors of modern salmon 25–100 mybp (Allendorf & Thorgaard 1984), in catostomids (suckers) about 50 mybp (Uyeno & Smith 1972), and in carp about 12 mybp (David et al. 2003). Partial duplications of the genome are probably more common, either as unequal exchanges during genetic recombination, or the unequal sorting of chromosomes during meiosis. This phenomenon can occur when two species hybridize, as observed in Pocilliiform fishes (guppies and mollies), occasionally giving rise to parthenogenic (unisexual) species.
Sex chromosomes
Gender in most vertebrates is determined by either chromosomes or hormones. In fishes both can occur. Most bony fishes and all elasmobranchs investigated to date have an XX/XY chromosome system, with males being the heterogametic (XY) sex (Maddock & Schwartz 1996; Devlin & Nagahama 2002). These include the diverse families Cichlidae, Gobiidae, Percidae, Fundulidae, Balistidae, and Salmonidae. A minority of fishes have the ZZ/ZW system, in which females are heterogametic. This arrangement dominates in some widespread families including Serranidae, Characidae, and Synodontidae. Both XY and ZW systems are found in the speciose families Cyprinidae and Poeciliidae, indicating that changes in sex determination systems can occur over relatively brief evolutionary timescales of a few million years (Devlin & Nagahama 2002). In contrast, the sex determination mechanisms in mammals (XY) and birds (ZW) are conserved across tens of millions of years (Mank et al. 2006).
As indicated by the many forms of hermaphroditism in fishes, gender defined by chromosomes can be altered by hormonal responses to environmental cues including temperature, season, and social status. For example, sex in the Atlantic Silverside (Menidia menidia) is determined by temperature; populations in cool weather can be 25% male, whereas the same population will be 50% male at the peak of summer (Conover & Kynard 1981). Overall, most fishes are gonochoristic (hard wired for one sex by chromosomes), but many others are flexible, and sex determination in these species may also be disrupted by organic pollutants that mimic sex hormones.
Mitochondrial genome
In the first decades of molecular genetic studies, the mitochondrial genome was much more accessible than the nuclear genome because it has more copies per cell, a small size, and is constructed like a single chromosome. Hence the study of mitochondrial genomics has proceeded much more rapidly than nuclear genomics, with over 100 complete fish mtDNA genomes resolved at this time (Miya et al. 2003). In contrast to the nuclear genome, the mtDNA has retained the same genes in the same locations and genome size is very similar across the vertebrates (Table 17.1). In fishes and most other vertebrates, the mtDNA genome encodes the information to construct 13 different proteins, 22 tRNAs, 2 rRNAs, and has a distinct segment for the origin of gene replication known as the control region.
Transgenic fishes
There is growing interest in genetically modified fishes to produce pharmaceutical products, novel aquarium pets, and faster growing strains for human consumption. This was originally attempted by “shotgunning”, injecting many copies of the desired gene into the nucleus of the eggs, a process with a very low success rate. Subsequent methods have grown more sophisticated, using viruses that can insert desired genes into a chromosome (Dunham 2004).
The first genetically modified fish was announced in 1984, based on Rainbow Trout (Oncorhynchus mykiss) eggs injected with a metallothionein (toxic metal resistance) gene (Maclean & Talwar 1984). Another early success was a transgenic Nile Tilapia (Oreochromis niloticus) modified with a gene for growth hormone. It attained three times the size of normal tilapia (Martinez et al. 1996). Rainbow Trout and Coho Salmon (Oncorhynchus kisutch) have also been modified with a salmon growth hormone gene that produced larger fish (Devlin et al. 2001). The first transgenic aquarium fish is the glofish®, a Zebrafish (Danio rerio) with red, green, and orange fluorescent colors (http://www.glofish.com/). This fluorescence gene may eventually be used to detect aquatic pollution by switching on a bright color in fishes that are exposed to environmental
contaminants.
Many authorities regard transgenic fishes as a potential boon to aquaculture, allowing higher survival, faster growing fishes, and larger yields. Indeed, genetically modified fishes have the potential to alleviate hunger and promote human health in impoverished corners of the globe. However, other authorities warn of the hazards of transgenic fishes escaping into the wild (Muir & Howard 2002). These hazards are similar to those of alien fish introductions, with the additional threat that transgenic fishes could breed with native stocks. In a study of a transgenic Medaka (Oryzias latipes), the modified males had greater mating success when introduced into a natural population, presumably because of their larger size, but their offspring had significantly lower survival. In these circumstances, accidental introduction of transgenic fishes might jeopardize future reproduction, possibly leading to extinction (Muir & Howard 1999). Clearly the benefits of this technology have to be balanced against significant risks.
Related Topics