Chapter: The Diversity of Fishes: Biology, Evolution, and Ecology: Fish genetics
Dispersal and vicariance revisited - Fish genetics: Phylogeography
Dispersal and vicariance revisited
As noted, modern biogeography has been dominated by the vicariance model, wherein species distributions are shaped by geographic isolation, rather than by active dispersal. In this framework, evolutionary history could be reconstructed due to breakthroughs in the study of plate tectonics (the movements of continents over millions of years). For the first time, the geographic distri butions of organisms could be interpreted through the sequence of continental breakups and collisions. While this emphasis on vicariance revitalized the field of biogeography, it was also dominated by a radical element that denied the primary alternative, dispersal (and colonization), as a means to explain species distributions. Vicariance biogeographers regarded dispersal as trivial or unprovable (de Queiroz 2005). In particular, the advocates for vicariance biogeography claimed that plate tectonics and other geological processes provided a testable set of expectations because they could be linked to geological events, whereas rare dispersal events did not readily fit into a hypothesis testing format. On the vicariance side of this debate, Nelson (1979) described dispersal biogeography as “a science of the improbable, the rare, the mysterious, and the miraculous”. In a courageous response to the radical view of vicariance, McDowall (1978) summarized the plight of adherents to dispersal theory:
How can one test for [dispersal] events that may occur once, a few, or even many times but which leave no trace of having occurred? One can’t. So we reach the point where, if we are going to insist on falsifiable theories, we must choose to exclude dispersal . . . but always we return to the fact that dispersal occurs.
Phylogeographic methods, and their immediate precursors in population genetics, provided a resolution to this dilemma. While it is true, as McDowall (1978) notes, that dispersal events are very difficult to document directly (especially in fishes), nonetheless these events yield clear genetic signals. This was dramatically demonstrated by Rosenblatt and Waples (1986), who used allozymes to test the prediction of an ancient vicariant separation between marine fishes of the East Pacific and central Pacific. The Pacific Ocean sits atop a geological plate that is over 100 million years old, and corresponding genetic divergences should be very deep. Instead, the allozymes revealed much more recent connections, on the order of thousands of years rather than millions of years. Lessios and Robertson (2006) revisited the issue 20 years later with mtDNA data, and found that 19 of 20 species either shared haplotypes across the barrier, or had haplotyes that were a few mutations apart. The exception was the pipefish Doryrhamphus excisus, a member of the Syngnathidae, known to have low
Although vicariance is an excellent model for many freshwater fishes, dispersal models are a better fit for marine fishes, given their large ranges and high potential for dispersal as both larvae and (in the case of tunas, billfishes, and pelagic sharks) swimming adults. However, there are exceptions to this trend, especially among marine fishes that lack a pelagic larval stage, like the Spiny Damselfish (Acanthochromis polyacanthus; Bay et al. 2006), Banggai Cardinalfish (Pterapogon kauderni; Hoffman et al. 2005) and seahorses (genus Hippocampus; Lourie etal. 2005). Vicariance models work well when population structure is shaped primarily by geographic barriers rather than life history or ecology. Accordingly, the next section will explore the phylogeography of freshwater fishes.
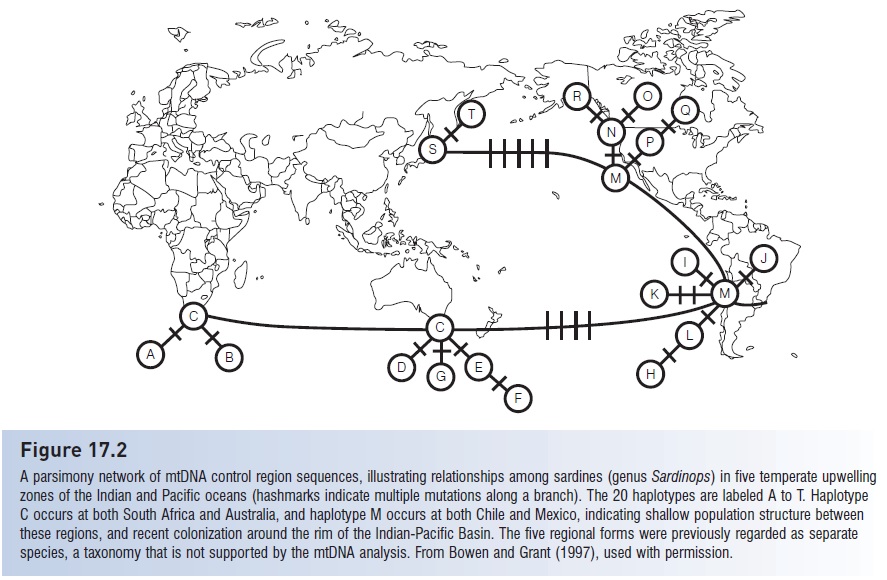
Figure 17.2
A parsimony network of mtDNA control region sequences, illustrating relationships among sardines (genus Sardinops) in five temperate upwelling zones of the Indian and Pacific oceans (hashmarks indicate multiple mutations along a branch). The 20 haplotypes are labeled A to T. Haplotype C occurs at both South Africa and Australia, and haplotype M occurs at both Chile and Mexico, indicating shallow population structure between these regions, and recent colonization around the rim of the Indian-Pacific Basin. The five regional forms were previously regarded as separate species, a taxonomy that is not supported by the mtDNA analysis. From Bowen and Grant (1997), used with permission.
Related Topics