Chapter: The Diversity of Fishes: Biology, Evolution, and Ecology: Fish genetics
Conservation genetics - Fish genetics
Conservation genetics
Genetics can contribute to conservation efforts in a number of ways. Molecular phylogenetic assessments can identify the oldest lineages in the tree of life, which contain a disproportionately high fraction of overall genetic diversity due to their age and uniqueness. Ancient lineages are not always obvious based on morphological examinations, as demonstrated in the bonefish case above. Treating multiple species as a single species would be a fundamentally flawed premise for fish management, and could put the less abundant species at risk. To serve the conservation goal of preserving biodiversity, we need to know the fundamental evolutionary lineages, both above and below the species level.
A second way that genetics can support conservation objectives is in defining populations, the fundamental units of wildlife management. The examples discussed previously illustrate how population genetics can assist these efforts. If two populations have significantly different allele frequencies, they are expected to bedemographically independent, meaning they have differences in demographic parameters such as age structure, fecundity, survivorship, growth rate, and sex ratio. However, for wildlife managers the more pragmatic concern is if an isolated population is depleted, it will not be replenished by dispersal from other populations. Isolated populations must recover from catastrophes, both natural and human-caused, without signifi - cant input of individuals from elsewhere. If the population goes extinct, the habitat may eventually be recolonized by rare migrants, but these colonists are not suffi cient to replenish populations over the timeframe of decades that concern wildlife managers.
Populations defined with genetics are often equated with stocks in fishery management, however they are not quite the same thing. If a group of fishes in one branch of a river is significantly different from elsewhere in terms of allele frequencies and F statistics, that genetically defined population can be regarded as an independent stock. However, the reverse is not always true. If fishes in two branches of the river are not significantly different in allele frequencies, then they may still be isolated stocks. It only takes a few migrants per generation to genetically homogenize breeding populations (but see Mills & Allendorf 1996). Ten effective migrants (meaning those that succeed in migrating to a new population and contributing genes to that population) may prevent population genetic differentiation, but will not be suffi cient to replenish depleted stocks. Hence a genetically isolated population is a stock, but a stock is not necessarily a genetically isolated population (Waples 1998). To assess contemporary movement and stock structure, tagging studies may be preferable. However, these are labor intensive, more expensive than most genetic assays, and impractical in many cases.
Fortunately this gap between demography and population genetics seems to be closing. New statistical methods allow researchers to identify individuals that move between populations by comparing microsatellite genotypes (Manel et al. 2005; Allendorf & Luikart 2006). Instead of relying on allele frequencies to define populations, these methods
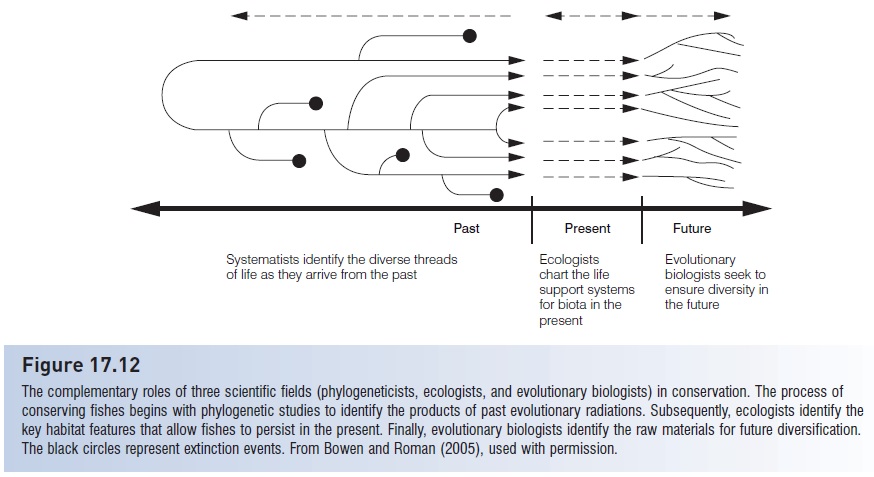
Figure 17.12
The complementary roles of three scientific fields (phylogeneticists, ecologists, and evolutionary biologists) in conservation. The process of conserving fishes begins with phylogenetic studies to identify the products of past evolutionary radiations. Subsequently, ecologists identify the key habitat features that allow fishes to persist in the present. Finally, evolutionary biologists identify the raw materials for future diversification. The black circles represent extinction events. From Bowen and Roman (2005), used with permission.
Hence microsatellites may close the gap between traditional population genetics, which assesses gene flow averaged across thousands of generations, and tagging studies that assess contemporary movement but may miss the rare or episodic exchanges.
The third major application of genetics in conservation is in the maintenance of evolutionary potential. This field has roots in the captive breeding programs that seek to retain genetic diversity and viability in endangered species (Frankham et al. 2002), and will draw on the emerging field of genomics. One goal is to preserve the genetic variants that will allow species to persist and survive future environmental challenges. This emphasis on adaptive evolutionary conservation (Fraser & Bernatchez 2001) can also include an assessment of novel genetic properties that confer selective advantages or higher survival, or could be the wellspring of new species.
Many technical challenges remain in the field of conservation genetics, such as developing the tools to read meaningful genetic changes among the millions of nucleotides that constitute a genome. Other challenges await in the realm of ethics and environmental responsibility. For example, when is it a good idea to clone endangered species? The first such clone was reported in 2000, a wild ox (Lanza et al. 2000); can fishes be far behind? Another challenge concerns the aquaculture of genetically modified fishes (Helfman 2007; see above, Fish genomics). Transgenic Zebrafish (Danio rerio) are now in routine development for commercial production of pharmaceuticals. What are the risks of using transgenic fish in aquaculture? Should we use transgenic technology to introduce pollution-resistant fishes? These ethical quandaries are right around the corner.
Molecular identification in the marketplace
The PCR technology that allows researchers to recover DNA data from small bits of tissue is now recognized as a major forensic tool, both in criminology and wildlife management. The first organized effort at species identification in the marketplace with PCR technology was directed at the Japanese and South Korean whale fisheries (Baker et al. 1996). To date the applications of forensic genetics to fish products have been few, but these cases are instructive.
Sturgeon caviar represents the ultimate luxury product from fishes, commanding prices upwards of US$50 per ounce. However, native stocks of the most prized species have crashed in the aftermath of the Soviet Union, as poorly regulated fisheries and high price have driven up the harvest, while pollution and dams have reduced habitat. In these circumstances, there is strong incentive to find substitutes for the premium caviar of the Volga River–Caspian Sea region. DeSalle and Birstein (1996) surveyed 23 lots of premium black caviar purchased from reputable dealers in New York City, using mtDNA sequences. They found that five of the lots (22%) were mislabeled eggs from less desirable but imperiled species, including three species listed on the Internation Union for the Conservation of Nature (IUCN) Red List (http://www.iucnredlist.org/) as Vulnerable (Siberian Sturgeon, Acipenser baerii) or Endangered (Amur River Sturgeon, A. schrenckii, and Ship Sturgeon, A. nudiventris).
Red Snapper (Lutjanus campechanus) is an esteemed fish in the restaurants and markets of North America, and commands a premium price. Yet few consumers have the discriminating pallet needed to be sure they are consuming the right species, and the genus Lutjanus has many members that are widespread, abundant, and delicious. In 1996 the Gulf of Mexico Fisheries Management Council imposed fishing restrictions after finding that the Red Snapper was overfished, driving down supply and driving up prices. Marko et al. (2004) surveyed specimens of Red Snapper purchased in eight states in the USA. The mtDNA cytochrome b sequences were compared to reference sequences available in Genbank (see above, Fish genomics). Seventeen of 22 specimens (77%) were not Red Snapper. Among the fraudulently labelled specimens, five were identified as other Atlantic snappers, two were Pacific Crimson Snapper (L. erythropterus), and the remaining 10 could not be identified because sequences from the corresponding species have not been submitted to Genbank. Some of these may be rare or unknown to science, invoking the possibility of overfishing before these species can be identified for management purposes. The fact that over half of the putative Red Snapper came from international sources indicates that this problem is global in scale.
Shark fin is one of the most contentious items in international wildlife trade, a commerce that takes an estimated 10 to 100 million sharks annually, and generates revenues equivalent to over a billion US dollars. In response to sharp declines in abundance worldwide, many countries have banned the practice of finning (harvesting the shark fins and discarding the rest of the fish), and three sharks (Whale, Basking, and Great White) are banned from international trade by the Convention on International Trade in Endangered Species. In these circumstances it is useful to know what species are entering the marketplace, and whether prohibited species are present. In response to this conservation concern, Shivji et al. (2002) developed diagnostic species-specific markers based on a nuclear ribosomal DNA sequence. In preliminary trials, 10 out of 55 putative Silky Sharks (Carcharhinus falciforis) proved to be other species. Subsequently Clarke et al. (2006) surveyed markets in Hong Kong and found that Blue Shark (Prionace glauca) predominated among auctioned fins (17%). Other sharks in the auctions included Shortfin Mako (Isurus oxyrinchus), Silky (C. falciformis), Sandbar (C. obscurus), Bull (C. leucas), hammerhead (Sphyrna spp.), and thresher (Alopias spp.).
These genetic surveys provide two lessons about the wildlife trade:
1 Legal markets such as those for Red Snapper in the USA are often a cover for poaching, smuggling, and illicit products entering the marketplace. Some of these products are from endangered or overutilized species.
2 Esteemed species are replaced by fraudulent alternatives. The practice of species mislabelling, dubbed “mock turtle syndrome”, is observed in 15–95% of luxury products surveyed to date, including caviar, fish fillets, shark fins, seal penises, whale meat, and turtle meat (Roman & Bowen 2000).
The response of wildlife management agencies to this illicit trade remains to be seen, but clearly the commerce in scarce fish products should be monitored. If you find some suspicious fish (or other wildlife) products, take a fin clip, a small tab of tissue, or a skin swab, and consult your local conservation geneticist.
Related Topics