Chapter: The Diversity of Fishes: Biology, Evolution, and Ecology: Fish genetics
Dispersal and population structure - Fish genetics
Dispersal and population structure
Certain life history traits correspond to shallow or deep population structure, especially those that influence the ability of the fish to disperse, as larva, juvenile, or adult. Hence the first generalization is that levels of population genetic structure are lowest in marine fishes, intermediate in anadromous fishes, and highest in freshwater fishes (Table 17.2). Sonoran topminnows (Poeciliopsis occidentalis) in the southwestern United States, that occupy desert springs separated by a few kilometers, can be isolated for thousands of years (Quattro et al. 1996). In this topminnow and other desert fishes, dispersal opportunities are limited to rare flooding events. At the other end of the spectrum, the Whale Shark (Rhincodon typus) has population structure only on the global scale of Atlantic versus Indo-Pacific oceans (Castro et al. 2007).
Genetic diversity (heterozygosity H) also shows a rank order among freshwater (lowest), anadromous (intermediate), and marine (highest) fishes. This is an expected consequence of tremendous differences in population size.
Freshwater populations may number in the thousands to millions, whereas their marine counterparts, with much larger ranges, may number in the millions to (in the case of anchovies and sardines) billions. Larger populations will accumulate more genetic diversity (Kimura 1983). There are many exceptions to these trends, but the conclusion of a rank order in genetic diversity is supported by both allozymes and microsatellite surveys (Table 17.2).
If populations are isolated for thousands of generations, they will eventually reach monophyly (see Deep population stucture entry above). The rate at which populations diverge depends on the effective population size (Ne), with a high probability of monophyly after 4Ne generations (Neigel & Avise 1986). Often the condition of monophyly is accompanied, upon closer examination, by morphological differences that indicate previously unrecognized cryptic species. However, this is not invariably the case, and scientists may prefer to retain a single taxonomic label that recognizes multiple evolutionary (subspecific) units within a species. The term evolutionary significant unit (ESU) was coined for subspecific evolutionary entities that show morphologiand
Table 17.2
Population genetic diversity averaged across three types of fishes, for allozymes (113 species; Ward et al. 1994) and for microsatellites (32 species; DeWoody & Avise 2000). Heterozygosity (H) values are progressively higher in freshwater, anadromous, and marine fishes. Population structure (FST) values from the allozyme survey are progressively lower in freshwater, anadromous, and marine fishes.
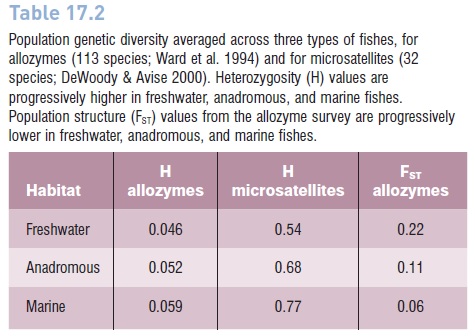
Table 17.3
Comparison of pelagic larval duration and population structure in 15 Atlantic reef fishes. Pelagic larval duration does not have a significant correlation with population structure (ΦST values). Surveys are based on mtDNA cytochrome b sequences except for the Pygmy Angelfish, which employed mtDNA control region sequences. The pelagic larval duration for Trumpetfish, Rock Hind, Soapfish, and Pygmy Angelfish are estimates from other members of the genus or family. An asterisk indicates species with deep population structure and suspected cryptic evolutionary lineages. From Bowen et al. (2006b).
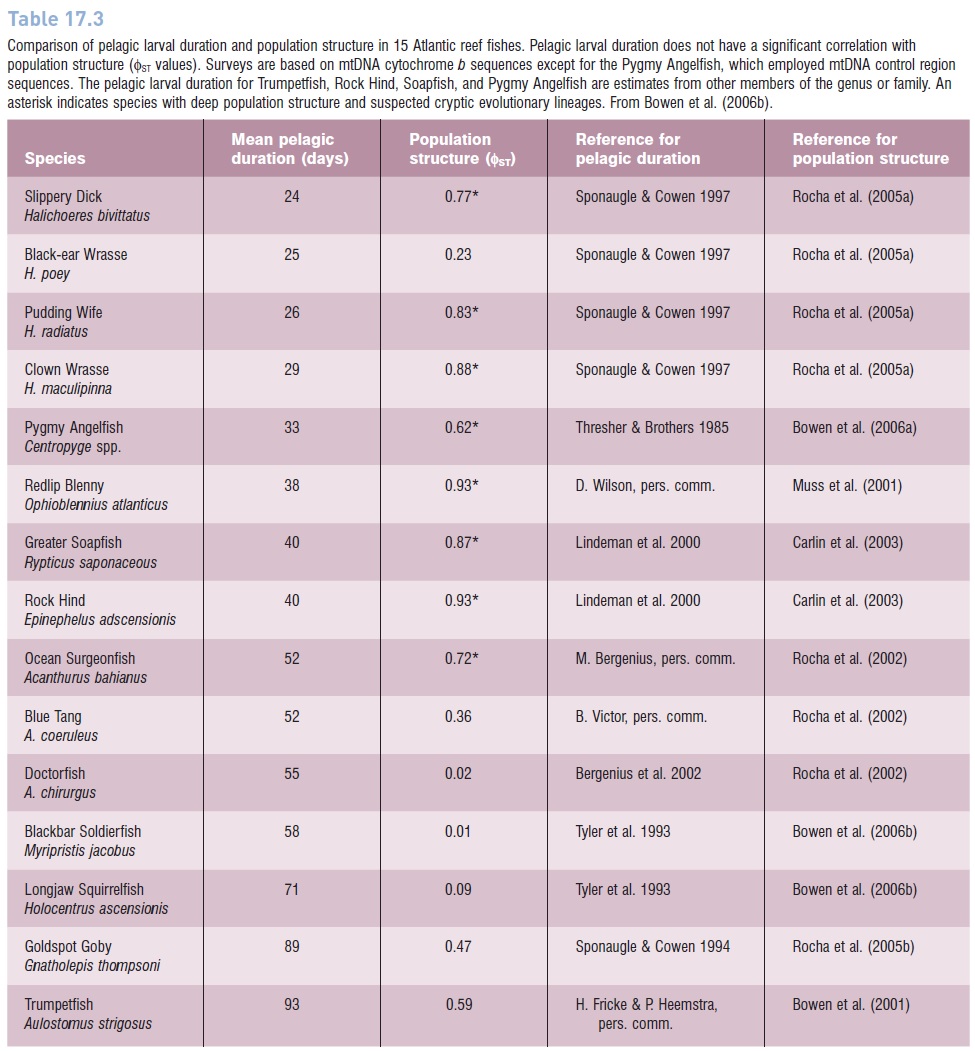
ESUs are often applied in the context of conservation, with an emphasis on higher priorities for ESUs than for populations, as has been applied to Pacific salmonids (see below). While monophyly in DNA assays is not the only way to assign such conservation priorities, this criterion is valuable for distinguishing populations that may have novel genetic characteristics, and may be in the process of speciating. ESUs as defined by monophyly of mtDNA sequences are surprisingly common in fishes, as indicated in Table 17.3, where eight out of 15 surveys of Atlantic reef fishes show evidence of ESUs.
Related Topics