Chapter: Pharmaceutical Biotechnology: Fundamentals and Applications : Pharmacokinetics and Pharmacodynamics of Peptide and Protein Drugs
Elimination of Protein Therapeutics - Pharmacokinetics of Protein Therapeutics
Elimination of Protein Therapeutics
Protein-based therapeutics are generally subject to the same catabolic pathways as endogenous or dietetic proteins. The end products of protein metabolism are thus amino acids that are reutilized in the endogenous amino acid pool for the de novo biosynthesis of structural or functional proteins in the human body (Meibohm, 2004). Detailed investigations on the metabolism of proteins are relatively difficult because of the myriad of potential molecule fragments that may be formed, and are therefore generally not conducted. Non-metabolic elimination pathways such as renal or biliary excretion are negligible for most proteins. If biliary excretion occurs, however, it is generally followed by subsequent metabolic degrada-tion of the compound in the gastrointestinal tract.
Proteolysis
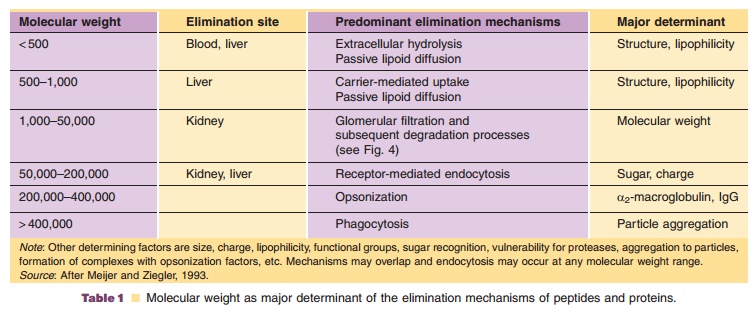
The metabolic rate for protein degradation generally increases with decreasing molecular weight from large to small proteins to peptides (Table 1), but is also dependent on other factors such as size, charge, lipophilicity, functional groups, and glycosylation pattern as well as secondary and tertiary structure.
The clearance of a peptide or protein describes the irreversible removal of active substance from the vascular space, which includes besides metabolism also cellular uptake. Proteolytic degradation of pro-teins can occur unspecifically nearly everywhere in the body or can be limited to a specific organ or tissue. Due to this unspecific proteolysis of some proteins already in blood as well as potential active cellular uptake, the clearance of protein drugs can exceed cardiac output, i.e., > 5 L/min for blood clearance and >3 L/min for plasma clearance (Meibohm, 2004).
Molecular weight determines the major metabo-lism site as well as the predominant degradation process (Wills, 1991). Proteolytic enzymes such as proteases and peptidases are ubiquitous throughout the body. Sites capable of extensive peptide and protein metabolism are not only limited to the liver, kidneys, and gastrointestinal tissue, but also include blood and vascular endothelium as well as other organs and tissues. As proteases and peptidases are also located within cells, intracellular uptake is per se more an elimination rather than a distribution process (Tang and Meibohm, 2006). While peptidases and proteases in the gastrointestinal tract and in lysosomes are relatively unspecific, soluble peptidases in the inter-stitial space and exopeptidases on the cell surface have a higher selectivity and determine the specific metabo-lism pattern of an organ. The proteolytic activity of SC tissue, for example, results in a partial loss of activity of SC compared to IV administrated interferon-g.
Gastrointestinal Protein Metabolism
As pointed out earlier, the gastrointestinal tract is a major site of protein metabolism with high proteolytic enzyme activity due to its primary function to digest dietary proteins. Thus, gastrointestinal metabolism of protein drugs is one of the major factors limiting systemic bioavailability of orally administered protein drugs. The metabolic activity of the gastrointestinal tract, however, is not limited to orally administered proteins. Parenterally administered peptides and pro-teins may also be metabolized in the intestinal mucosa following intestinal secretion. At least 20% of the degradation of endogenous albumin, for example, has been reported to take place in the gastrointestinal tract (Colburn, 1991).
Renal Protein Metabolism and Excretion
The kidneys are a major site of protein metabolism for smaller sized proteins that undergo glomerular filtration. The size-selective cut-off for glomerular filtration is approximately 60 kD, although the effec-tive molecule radius based on molecular weight andconformation is probably the limiting factor (Edwards et al., 1999). Glomerular filtration is most efficient, however, for proteins smaller than 30 kDa (Kompella and Lee, 1991). Peptides and small proteins (< 5 kDa) are filtered very efficiently, and their glomerular filtration clearance approaches the glomerular filtra-tion rate (GFR, ~120 mL/min in humans). For mole-cular weights exceeding 30 kDa, the filtration rate falls off sharply. In addition to size selectivity, charge selectivity has also been observed for glomerular filtration where anionic macromolecules pass through the capillary wall less readily than neutral macro-molecules, which in turn pass through less readily than cationic macromolecules (Deen et al., 2001).
The importance of the kidneys as elimination organ could for example be shown for interleukin-2, macrophage-colony-stimulating factor (M-CSF) and interferon-a (McMartin, 1992; Wills and Ferraiolo, 1992). The relative contributions of renal and hepatic clearances to the total plasma clearance of several proteins are shown in Figure 3.Renal metabolism of peptides and small proteins is mediated through three highly effective processes (Fig. 4). As a result, only minuscule amounts of intact protein are detectable in urine.
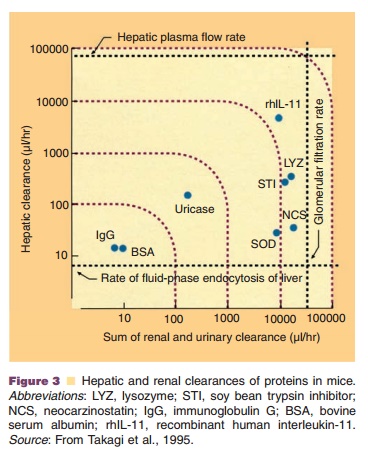
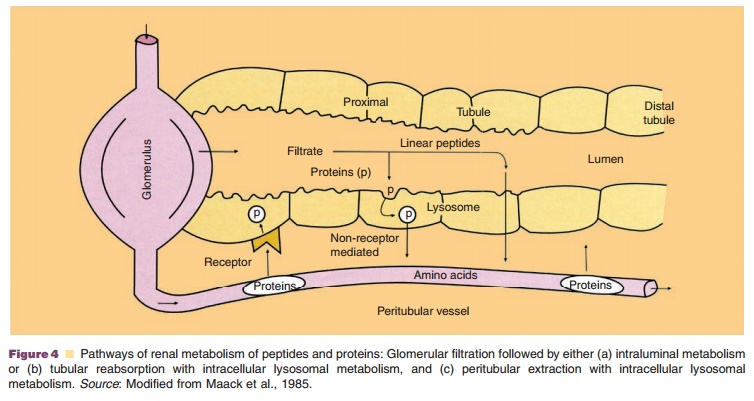
The first mechanism involves glomerular filtra-tion of larger, complex peptides and proteins followed by reabsorption into endocytic vesicles in the prox-imal tubule and subsequent hydrolysis into small peptide fragments and amino acids (Maack et al., 1985). This mechanism of elimination has been described for IL-2 (Anderson and Sorenson, 1994), IL-11 (Takagi et al., 1995), growth hormone (Johnson and Maack, 1977), and insulin (Rabkin et al., 1984).
The second mechanism entails glomerular filtra-tion followed by intraluminal metabolism, predomi-nantly by exopeptidases in the luminal brush border membrane of the proximal tubule. The resulting peptide fragments and amino acids are reabsorbed into the systemic circulation. This route of disposition applies to small linear peptides such as glucagon and LH-RH (Carone and Peterson, 1980; Carone et al., 1982). Recent studies implicate the proton driven peptide transporters PEPT1 and especially PEPT2 as the main route of cellular uptake of small peptides and peptide-like drugs from the glomerular filtrates (Inui et al., 2000). These high-affinity transport proteins seem to exhibit selective uptake of di- and tripeptides, which implicates their role in renal amino acid homeostasis (Daniel and Herget, 1997).
For both mechanisms, glomerular filtration is the dominant, rate-limiting step as subsequent degrada-tion processes are not saturable under physiologic conditions (Maack et al., 1985; Colburn, 1991). Due to this limitation of renal elimination, the renal contribution to the overall elimination of proteins is dependent on the proteolytic activity for these proteinsin other body regions. If metabolic activity for theseproteins is high in other body regions, there is onlyminor renal contribution to total clearance, and itbecomes negligible in the presence of unspecific degradation throughout the body. If the metabolic activity is low in other tissues or if distribution to the extravascular space is limited, however, the renal contribution to total clearance may approach 100%. This is for instance the case for recombinant human interleukin-10 (rhIL-10), for which clearance correlates closely with GFR, making dosage adjustments neces-sary in patients with impaired renal function (Andersen et al., 1999).
The third mechanism of renal metabolism is peritubular extraction of peptides and proteins from post-glomerular capillaries with subsequent intracel-lular metabolism. Experiments using radio-iodinated growth hormone (125I-rGH) have demonstrated that while reabsorption into endocytic vesicles at the proximal tubule is still the dominant route of disposition, a small percentage of the hormone may be extracted from the peritubular capillaries (Johnson and Maack, 1977; Krogsgaard Thomsen et al., 1994). Peritubular transport of proteins and peptides from the basolateral membrane has also been shown for insulin (Nielsen et al., 1987).
Hepatic Protein Metabolism
Aside from renal and gastrointestinal metabolism, theliver may also play a major role in the metabolism ofprotein therapeutics. Exogenous as well as endogenousproteins undergo proteolytic degradation to dipeptidesand amino acids that are reused for endogenous protein synthesis. Proteolysis usually starts with endopepti-dases that attack in the middle part of the protein, and the resulting oligopeptides are then further degraded by exopeptidases. The rate of hepatic metabolism is largely dependent on the specific amino acid sequence of the protein (Meibohm, 2004).
A prerequisite for hepatic protein metabolism is the uptake of proteins into the hepatocytes. An overview of the different mechanisms of hepatic uptake of proteins is listed in Table 2.
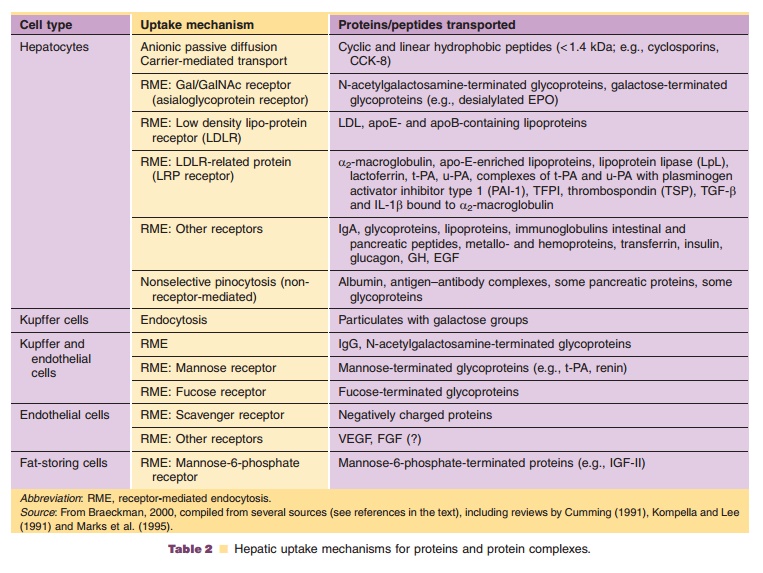
Small peptides may cross the hepatocyte mem-brane via simple passive diffusion if they have sufficient hydrophobicity. Peptides of this nature include the cyclosporins (cyclic peptides) (Ziegler et al., 1988). Other cyclic and linear peptides of small size (< 1.4 kDa) and hydrophobic nature (containing aromatic amino acids), such as cholecystokinin-8 (CCK-8; 8 amino acids), are taken up by the hepatocytes by a carrier-mediated transport (Ziegler et al., 1988), in the case of CCK-8 by the organic anion transporting polypeptide OATP-8 (SLCO1B3) (Ismair et al., 2001). After internalization into the cytosol,
these peptides are usually metabolized by microsomal enzymes (cytochrome P-450 3A for cyclosporin A) or cytosolic peptidases (CCK-8). Substances that enter the liver via carrier-mediated transport are typically excreted into the bile by active export transporters. These hepatic clearance pathways are identical to those known for most small organic hydrophobic drug molecules.
Uptake of larger peptides and proteins is facilitated via various carrier-mediated, energy-dependent transport processes. One of the possibili-ties is receptor-mediated endocytosis, such as for insulin and epidermal growth factor (Kim et al., 1988; Sugiyama and Hanano, 1989; Burwen and Jones, 1990). In receptor-mediated endocytosis, circulating proteins are recognized by specific hepatic receptor proteins (Kompella and Lee, 1991). The receptors are usually integral membrane glycoproteins with an exposed binding domain on the extracellular side of the cell membrane. After the binding of the circulating protein to the receptor, the complex is already present or moves in coated pit regions, and the membrane invaginates and pinches off to form an endocytotic coated vesicle that contains the receptor and ligand (internalization). The vesicle coat consists of proteins (clathrin, adaptin, and others), which are then removed by an uncoating adenosine triphosphatase (ATPase). The vesicle parts, the receptor, and the ligand dissociate and are targeted to various intracel-lular locations. Some receptors, such as the low-density lipoprotein (LDL), asialoglycoprotein and transferrin receptors, are known to undergo recycling. Since sometimes several hundred cycles are part of a single receptor’s lifetime, the associated receptor-mediated endocytosis is of high capacity. Other receptors, such as the interferon receptor, undergo degradation. This degradation leads to a decrease in the concentration of receptors on the cell surface (receptor down-regulation). Others, such as insulin receptors, for example, undergo both recycling and degradation (Kompella and Lee, 1991).
For glycoproteins, if a critical number of exposed sugar groups (mannose, galactose, fucose, N-acetyl-glucosamine, N-acetylgalactosamine, or glucose) is exceeded, receptor-mediated endocytosis through sugar-recognizing receptors is an efficient hepatic uptake mechanism (Meijer and Ziegler, 1993). Important carbohydrate receptors in the liver are the asialoglycoprotein receptor in hepatocytes and the mannose receptor in Kupffer and liver endothelial cells (Smedsrod and Einarsson, 1990; Bu et al., 1992). The high-mannose glycans in the first kringle domain of t-PA, for example, have been implicated in its hepatic clearance (Cumming, 1991).
Low density lipoprotein receptor-related protein (LRP) is a member of the LDL receptor family responsible for endocytosis of several important lipo-proteins, proteases, and protease-inhibitor complexes in the liver and other tissues (Strickland et al., 1995). Examples of proteins and protein complexes for which hepatic uptake is mediated by LRP are listed in Table 2.
Uptake of proteins by liver cells is followed by transport to an intracellular compartment for meta-bolism. Proteins internalized into vesicles via an endocytotic mechanism undergo intracellular trans-port towards the lysosomal compartment near the center of the cell. There, the endocytotic vehicles fuse with or mature into lysosomes, which are specialized acidic vesicles that contain a wide variety of hydro-lases capable of degrading all biological macromole-cules. Proteolysis is started by endopeptidases (mainly cathepsin D) that act on the middle part of the proteins. Oligopeptides—as the result of the first step—are further degraded by exopeptidases. The resulting amino acids and dipeptides reenter the metabolic pool of the cell (Meijer and Ziegler, 1993). The hepatic metabolism of glycoproteins may occur more slowly than the naked protein becauseprotecting oligosaccharide chains need to be removed first. Metabolized proteins and peptides in lysosomes from hepatocytes, hepatic sinusoidal cells and Kupffer cells may be released into the blood. Degraded proteins in hepatocyte lysosomes can also be deliv-ered to the bile canaliculus and excreted by exocytosis.
A second intracellular pathway for proteins is the direct shuttle or transcytotic pathway (Kompella and Lee, 1991). The endocytotic vesicle formed at the cell surface traverses the cell to the peribiliary space, where it fuses with the bile canalicular membrane, releasing its contents by exocytosis into bile. This pathway, described for polymeric immunoglobulin A, bypasses the lysosomal compartment completely.
Receptor-Mediated Protein Metabolism
Receptor-mediated metabolism is often a substantial elimination pathway for those protein therapeutics that bind with high affinity to membrane-associated receptors on the cell surface. The interaction of the protein therapeutic with the membrane receptor is frequently part of the pharmacologic effect of the drug, i.e., the receptor is the target structure the protein therapeutic is directed at. This binding can lead to receptor-mediated uptake by endocytosis and subsequent intracellular lysosomal metabolism. Receptor-mediated uptake and metabolism via inter-action with these generally high-affinity, low-capacity binding sites is not limited to a specific organ or tissue type. Thus, any tissue, including the therapeutic target cells that express receptors for the drug can contribute to the elimination of the protein therapeu-tic (Meibohm and Derendorf, 2003).
Since the number of protein drug receptors is limited, receptor-mediated protein metabolism can usually be saturated within therapeutic concentra-tions, or more specifically at relatively low molar ratios between the protein drug and the receptor. As a consequence, the elimination clearance of these protein drugs is not constant but dose dependent, and decreases with increasing dose. Thus, receptor-mediated elimination constitutes a major source for nonlinear pharmacokinetic behavior of numerous peptide and protein drugs, i.e., systemic exposure to the drug increases more than proportional with increasing dose (Tang et al., 2004).
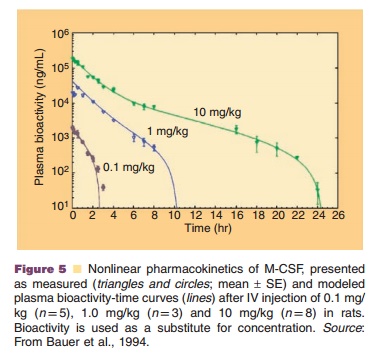
Recombinant human M-CSF, for example, un-dergoes besides linear renal elimination a nonlinear elimination pathway that follows Michaelis–Menten kinetics and is linked to a receptor-mediated uptake into macrophages. At low concentrations, M-CSF follows linear pharmacokinetics, while at high con-centrations nonrenal elimination pathways are satu-rated resulting in nonlinear pharmacokinetic behavior (Fig. 5) (Bartocci et al., 1987; Bauer et al., 1994).
Related Topics