Chapter: Genetics and Molecular Biology: Lambda Phage Integration and Excision
Structure of the att Regions
Structure of the att Regions
Historically the ability to sequence DNA came after
geneticists had isolated the att
mutants and had determined much about the integration

Figure
18.16 Theattregions andnucleotide sequence of the O region of att.
Therefore, obtaining the
sequence of att would permit checking
the genetic inferences. Sequencing the att
region was straightforward because phage carrying the four possible att regions— POP’, BOB’, BOP’, and POB’—were available. Landy identified the ap-propriate restriction
fragments from these phage, isolated and sequenced them. As expected from the
genetic experiments, all four contained an identical central or core sequence
corresponding to the O part of the att region. The size of the core could
not have been deter-mined from the genetic experiments, but it turned out to be
15 base pairs long (Fig. 18.16).
The recognition regions flanking the cores are of
different lengths. By deletion analysis, the sequences on either side of O at the bacterial att region that are necessary for integration have been shown to be
justlarge enough to specify an Int protein binding site. By contrast, attP is about 250 base pairs long.
The sites of Int-promoted cleavage of the O sites have been deter-mined exactly
using radioactive phosphate both as a label and as an agent to cleave the DNA.
An in vitro integration reaction was
performed between an attP and a short
DNA fragment containing attB that had
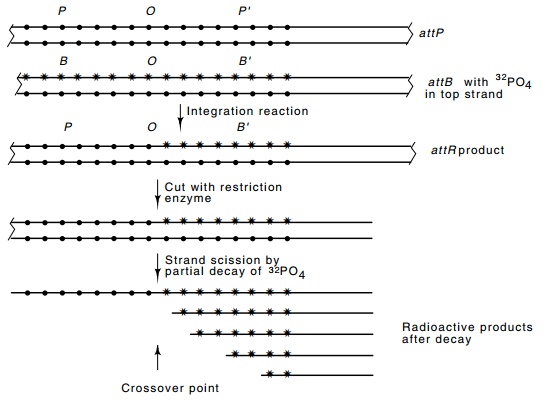
Figure 18.17 Scheme for determining the site of strand transfer in the inte-gration reaction. Small circles represent nonradioactive phosphates and stars represent radioactive phosphates. More precision is gained by placing radioac-tive phosphates adjacent to only one of the four bases by using the appropriate α-labeled triphosphates when the strand is labeled.
been extensively labeled with radioactive phosphate
on one and then the other strand. This generated attL and attR sites. In
the attR site, all phosphates that
lie to the right of the crossover point derive from attB and are therefore both radioactive and subject to strand
scission by radioactive decay. The positions of these radioactive phosphates
were then determined by permitting some to decay. The decay event cleaves the
phosphate backbones; subsequent electrophoresis of the denatured fragments on a
sequencing gel yields bands corresponding to each position that had a radioactive
phosphate and no bands of other sizes (Fig. 18.17). The exchange points are
deduced to be at fixed positions on the top and bottom strands, but separated
by seven base pairs.
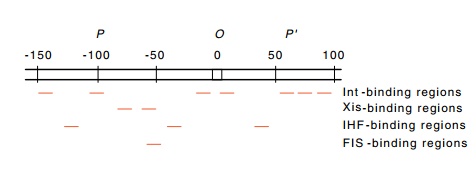
Figure 18.18 The regions inattPto which Int, Xis, FIS, and IHF proteins bind.
Related Topics