Chapter: Genetics and Molecular Biology: Protein Structure
Locating Specific Residue-Base Interactions - Protein Structure
Locating Specific Residue-Base Interactions
We discussed how a histone bound to DNA can protect
the nucleotides it contacts from DNAse cleavage of phosphodiester bonds.
Biochemical methods similar to those DNAse footprinting experiments can even
identify interactions between specific amino acid residues and specific bases.
In some cases the biochemical approaches provide as much resolution as X-ray
crystallography.
First, consider how to test a guess of a specific
residue-base interac-tion. In this approach a correct guess can be confirmed,
but a poor guess yields little information. The idea is that if the amino acid
residue in question is substituted by a smaller residue, say a glycine or
alanine,
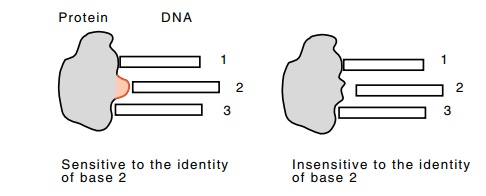
Figure
6.20 How replacing a DNA contacting
residue of a protein with asmaller residue like alanine or glycine makes the
protein indifferent to the identity of the base that formerly was contacted by
the residue.
then the former interaction between the residue and
the base will be eliminated. As a result, the strength of the protein’s binding
becomes independent of the identity of the base formerly contacted, whereas the
wild-type unsubstituted protein would vary in its affinity for the DNA as the
contacted base is varied (Fig. 6.20).
The work required for this missing contact
experiment is high. Genetic engineering techniques must be used to alter the
gene encoding the protein. Similarly, both the wild-type sequence and a
sequence for each base being tested for contacting must also be synthesized.
Finally, the protein must be synthesized in
vivo, purified, and its affinity for the various DNAs tested.
The testing for specific contacts can be
streamlined. Instead of having to guess correctly both the base and the
residue, the technique finds whatever base is contacted by a residue. As
before, a variant protein is made containing a glycine or alanine substitution
in the residue sus-pected of contacting the DNA. The DNA sample is probed
biochemically to find all the residues contacted by the wild-type protein and
by the mutant protein. The difference between the set of bases contacted by the
wild-type protein and the mutant protein is the base or two that are normally
contacted by the altered residue. The procedure for locating the bases which contact
the protein is more fully described.
Related Topics