Chapter: Genetics and Molecular Biology: Protein Structure
Electrostatic Forces that Determine Protein Structure - Protein Structure
Electrostatic Forces that Determine Protein
Structure
Proteins have a difficult time surviving. If we
heat them a little above the temperatures normally found in the cells from
which they are isolated, most are denatured. Why should this be? On first
consideration it would make sense for proteins to be particularly stable and to
be able to withstand certain environmental insults like mild heating. One
expla-nation for the instability is that proteins just cannot be made more
stable. A second possibility is that the instability is an inherent part of
proteins’ activities. This latter possibility seems more likely since en-zymes
extracted from bacteria that thrive at temperatures near the boiling point of
water frequently are inactive at temperatures below 40°. It could be that to
act as catalysts in chemical reactions or to participate in other cellular
activities, proteins must be flexible, and such flexibility means that proteins
must exist on the verge of denaturing. A final possibility is that rapid
fluctuations in structure of the folding interme-diates are necessary for a
protein to find the correct folded conforma-tion. The existence of such
meta-stable states may preclude the existence of a highly stable folded state.
Future research should illuminate this
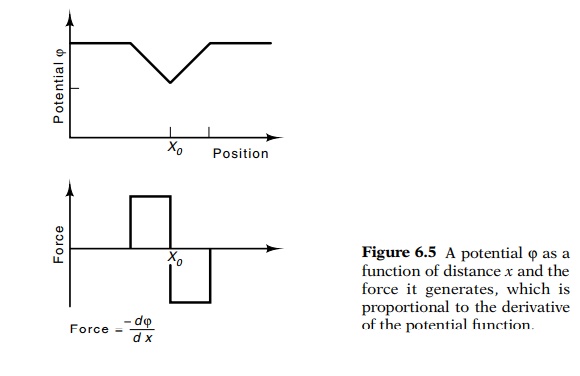
Often it is helpful to think of the interactions
between amino acids in terms of forces. In some cases, physicists and physical
chemists have also found it convenient to consider the interactions between
objects in terms of potentials (Fig. 6.5). Some of our discussion will be more
streamlined if we, also, use potentials. Forces and potentials are easily
interconvertible since forces are simply related to potentials. The steep-ness
of an object’s potential at a point is proportional to the force on the object
while at that point,
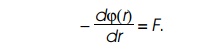
Alternatively, the potential difference between two
points is propor-tional to the work required to move an object between the two
points.
Figure
6.6 The electrostatic force between
two charges of valueQ1andQ2separated by a distance r and the potential ϕ produced
by a single charge Q1.
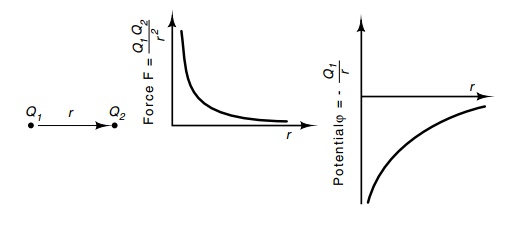
Electrostatics are the basis of several of the
forces that determine protein structure. Charges of unlike sign attract each
other with a force proportional to each of the charges and inversely
proportional to the square of the charge separation (Fig. 6.6). The force is
also inversely proportional to the dielectric constant of the medium, but for
purposes of our discussion, this factor will not be considered. Thus, the
potential generated at a point by a charge is proportional to the magnitude of
the charge and inversely proportional to the distance of the point from the
charge
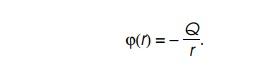
The electric field at a point is defined as the
force a unit charge would feel at that point. Therefore, the magnitude of the
electric field at a point r generated
by a charge Q at the origin is
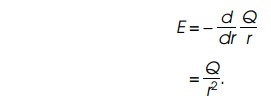
Since both positively and negatively charged amino
acids exist, direct electrostatic attractions are possible in proteins. These
generate what are called salt bridges. A number of proteins contain such salt
bridges.
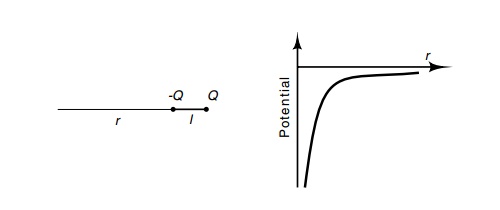
When two equal and opposite charges or partial
charges are located near one another, as is found in the polar amino acids and
in each peptide bond, it is convenient to view their combined effect on other
atoms and molecules as a whole rather than considering each charge
individually. A dipole with charges +Q
and -Q separated by a distance l (Fig. 6.7) generates a potential and
electric field proportional to
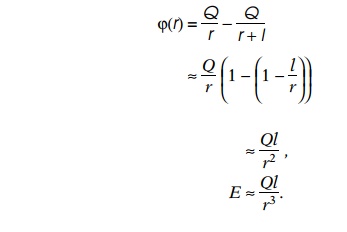
Interactions
between pairs of dipoles are also common in proteins. By the same reasoning as
used above, the potential between the two dipoles ql and QL can be shown to
be proportional to
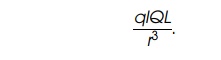
Even more important to protein structure and
function than the interactions between permanent dipoles in proteins are the
momentary interactions between temporary dipoles that have been created by a
brief fluctuation in the positions of charges. The forces generated by
interactions between such dipoles are called London dispersion forces. They are
weak, short-ranged, up to an Angstrom or two, attractive forces that exist
between all molecules. These form the basis of most of the selectivity in the
binding of other molecules to proteins. If the shapes of a protein and another
molecule are complementary, then many of these attractive forces can act and
hold the two molecules tightly together. If
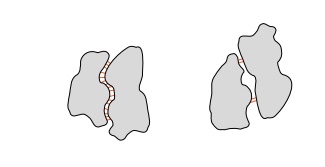
the shapes are not exactly complementary, then
because the forces are short-ranged, only the small areas in contact are
subject to dispersion attractive forces and the two molecules do not strongly
bind to one another.
Dispersion forces are particularly short-ranged
since their attractive potential falls off with the sixth power of the distance
separating the molecules. Although these forces are best understood in the
framework of quantum mechanics, we can understand the origin of the sixth-power
dependence. Consider an electrically neutral nonpolar molecule. Thermal
fluctuations can generate a momentary separation of its plus and
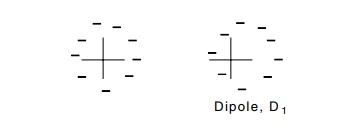
minus charges. That is, a dipole of strength D1 = ql is briefly generated. The electric field produced by this dipole can induce a dipole in an adjacent susceptible molecule. The strength of the induced dipole is directly proportional to the strength of the local electric field. That is, the induced dipole has a strength, D2, proportional to

Since, as discussed above, the potential between
two dipoles is proportional to the product of their strength and inversely
proportional to the cube of their separation,
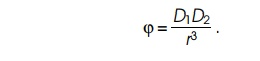
Substituting the value for D2 in the potential yields the result that the potential between the two
dipoles is inversely proportional to the sixth power of their separation,
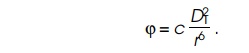
Due to the inverse sixth-power dependence, the
dispersion forces become much stronger as the distance separating two molecules
be-comes smaller. The force cannot become too great, for once the elec-tronic
cloud of one molecule begins to interpenetrate the cloud of the other molecule,
a very strong repulsive interaction sets in. It is compu - tationally
convenient to approximate this repulsive potential as an inverse twelfth power
of the separation of the centers of the atoms. The combination of the two
potentials is known as a Van der Waals potential. The radius at which the
strong repulsion begins to be significant is the Van der Waals radius (Fig.
6.8).
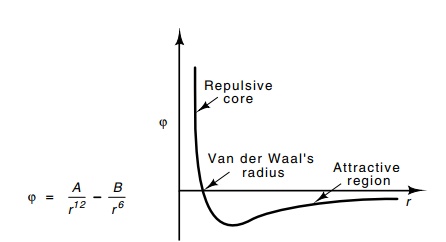
Figure 6.8 A Van der Waals potential.
Related Topics