Biotechnology - Tools for Genetic Engineering | 12th Botany : Chapter 4 : Principles and Processes of Biotechnology
Chapter: 12th Botany : Chapter 4 : Principles and Processes of Biotechnology
Tools for Genetic Engineering
Tools for Genetic Engineering
Now we know from the foregoing discussion that in order to
generate recombinant DNA molecule, certain basic tools are necessary for the
process. The basic tools are enzymes, vectors and host organisms. The most
important enzymes required forgenetic engineering are the restriction
enzymes, DNA ligase and alkaline phosphatase.
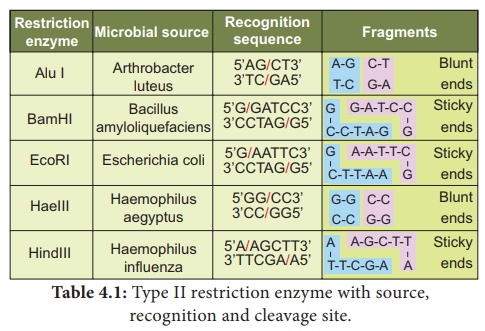
1. Restriction Enzymes
The two enzymes responsible for restricting the growth of
bacteriophage in Escherichia coli were isolated in the year 1963. One was the
enzyme which added methyl groups to DNA,while the other cut DNA. The later was
called restriction endonuclease. A restriction enzyme or restriction
endonuclease is an enzyme that cleaves DNA into fragments at or near specific
recognition sites within the molecule known as restriction sites. Based on
their mode of action restriction enzymes are classified into Exonucleases and
Endonucleases.
a. Exonucleases are enzymes which remove nucleotides one at a time
from the end of a DNA molecule. e.g. Bal 31, Exonuclease III.
b.Endonucleases are enzymes which break the internal
phosphodiester bonds within a DNA molecule. e.g. Hind II, EcoRI, Pvul, BamHI,
TaqI.
Restriction endonuclease: Molecular scissors
The restriction enzymes are called as molecular scissors. These
act as foundation of recombinant DNA technology. These enzymes exist in
many bacteria where they function as a part of their defence mechanism called
restriction- modification system.
There are three main classes of restriction endonuclease : Type I,
Type II and Type III, which differ slightly by their mode of action. Only type
II enzyme is preferred for use in recombinant DNA technology as they recognise
and cut DNA within
a specific sequence typically consisting of 4-8 bp.
Examples of certain enzymes are given in table 5.1.
The restriction enzyme Hind II always cut DNA molecules at a point
of recognising a specific sequence of six base pairs. This sequence is known s
recognition sequence. Today more than 900 restriction enzymes that
have been isolated from over 230 strains of bacteria with different recognition
sequences.
Restriction endonucleases are named by a standard procedure. The
first letter of the enzymes indicates the genus name, followed by the first two
letters of the species, then comes the strain of the organism and finally a
roman numeral indicating the order of discovery. For example, EcoRI is from
Escherichia (E) coli (co), strain RY 13 (R) and first
endonuclease (I) to be discovered.
It contains 2 different antibiotic resistance genes and
recognition site for several restriction enzymes. This sequence is referred to
as a restriction site and is generally –palindromic which means that the sequence
in both DNA strands at this site read same in 5’ – 3’ direction and in the
3’-5’ direction
Example: MALAYALAM: This phrase is read the same in either of the
directions.
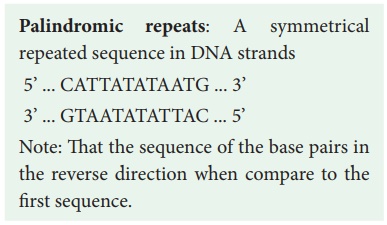
Palindromic repeats: A symmetrical
repeated sequence in DNA strands
5’ ... CATTATATAATG ...
3’
3’ ... GTAATATATTAC ...
5’
Note: That the sequence of the base pairs in the reverse
direction when compare to the first sequence.
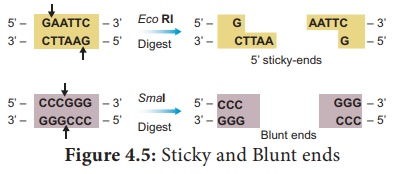
The exact kind of cleavage produced by a restriction enzyme is
important in the design of a gene cloning experiment. Some cleave both strands
of DNA through the centre resulting in blunt or flush end. These
are known as symmetric cuts. Some enzymes cut in a way producing protruding and
recessed ends known as sticky or cohesive end. Such cut are
called staggered or asymmetric cuts.
Two other enzymes that play an important role in recombinant DNA
technology are DNA ligase and alkaline phosphatase
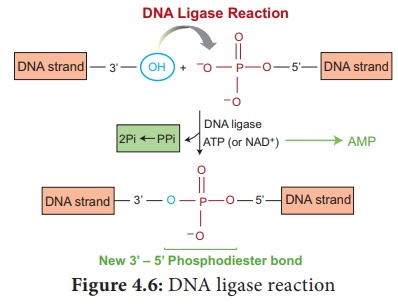
2. DNA Ligase
DNA ligase enzyme joins the sugar and phosphate molecules of double stranded DNA (dsDNA) with 5’-PO4 and a 3’-OH in an Adenosine Triphosphate (ATP) dependent reaction. This is isolated from T4 phage.
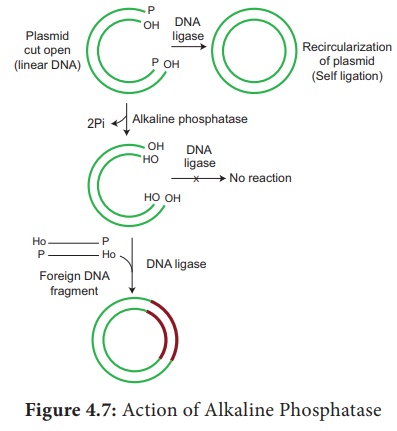
3. Alkaline Phosphatase
It is a DNA modifying enzymes and adds or removes specific
phosphate group at 5’ terminus of double stranded DNA (dsDNA) or single
stranded DNA (ssDNA) or RNA. Thus it prevents self ligation. This enzyme is
purified from bacteria and calf intestine.
4. Vectors
Another major component of a gene cloning experiment is a vector
such as a plasmid. A Vector is a small DNA molecule capable of self-replication
and is used as a carrier and transporter of DNA fragment which is inserted
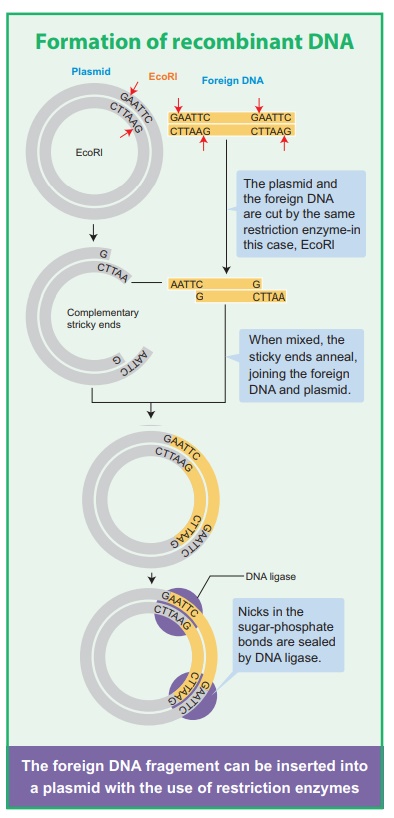
Vector is also called cloning
vehicle or cloning DNA. Vectors are of two types: i) Cloning Vector,
and
ii)Expression Vector. Cloning vector is used for the cloning of
DNA insert inside the suitable host cell. Expression vector is used to express
the DNA insert for producing specific protein inside the host.
Properties of Vectors
Vectors are able to replicate autonomously to produce multiple
copies of them along with their DNA insert in the host cell.
·
It should be small in size and of low molecular weight, less than
10 Kb (kilo base pair) in size so that entry/transfer into host cell is easy.
·
Vector must contain an origin of replication so that it can
independetly replicate within the host.
·
It should contain a suitable marker such as antibiotic resistance,
to permit its detection in transformed host cell.
·
Vector should have unique target sites for integration with DNA
insert and should have the ability to integrate with DNA insert it carries into
the genome of the host cell. Most of the commonly used cloning vectors have
more than one restriction site. These are Multiple Cloning Site (MCS) or
polylinker. Presence of MCS facilitates the use of restriction enzyme of
choice.
The following are the features that are required to
facilitate cloning into a vector.
1. Origin of replication (ori): This is a sequence from where replication
starts and piece of DNA when linked to this sequence can be made to replicate
within the host cells.
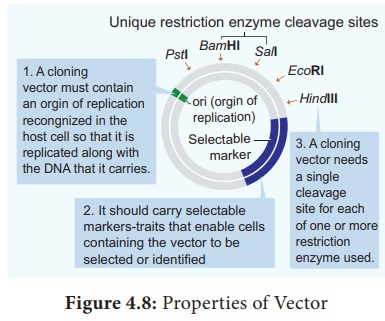
2. Selectable marker: In addition to ori the vector
requires a selectable marker, which helps in identifying and eliminating non
transformants and selectively permitting the growth of the transformants.
3. Cloning sites: In order to link the alien DNA, the
vector needs to have very few, preferably single, recognition sites for the
commonly used restriction enzymes.
Types of vector
Few types of vectors are discussed in detail below:
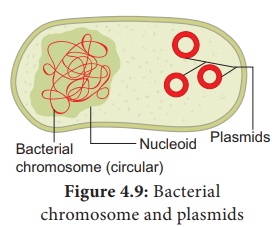
Plasmid
Plasmids are extra c h rom o s om a l , self replicating ds
circular DNA molecules, found in the bacterial cells in addition to the
bacterial chromosome. Plasmids contain Genetic information for their own
replication.
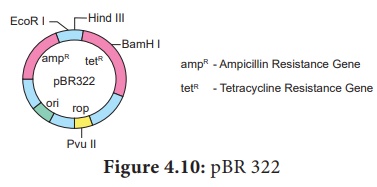
pBR 322 Plasmid
pBR 322 plasmid is a reconstructed plasmid and most widely used as
cloning vector; it contains 4361 base pairs. In pBR, p denotes
plasmid, Band R respectively the names of scientist
Boliver and Rodriguez who developed this plasmid. The number 322
is the number of plasmid developed from their laboratory. It contains ampR and
tetR two different antibiotic resistance genes and recognition sites for
several restriction enzymes. (Hind III, EcoRI, BamH I, Sal
I, Pvu II, Pst I, Cla I), ori and
antibiotic resistance genes. Rop codes for the proteins involved in the
replication of the plasmid.
Ti Plasmid
Ti plasmid is found in Agrobacterium tumefaciens, a bacteria
responsible for inducing tumours in several dicot plants. The plasmid carries
transfer(tra) gene which help to transfer
T- DNA from one bacterium to other bacterial or plant cell.
It has Onc gene for oncogenecity,
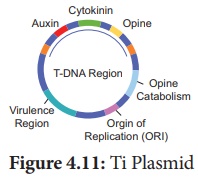
ori gene for origin for replication and inc gene for
incompatibility. T-DNA of Ti-Plasmid is stably integrated with plant DNA.
Agrobacterium plasmids have been used for introduction of genes of desirable
traits into plants.
Transposon as Vector
Transposons (Transposable elements or mobile elements) are DNA
sequence able to insert itself at a new location in the genome without having
any sequence relationship with the target locus and hence transposons are
called walking genes or jumping genes. They are used as
genetic tools for analysis of gene and protein functions, that produce new
phenotype on host cell.The use of transposons is well studied in Arabidopsis
thaliana and bacteria such as Escherichia coli.

Walking Genes - Gene walking
involves the complete sequencing of
large more than 1 kb stretches of DNA.
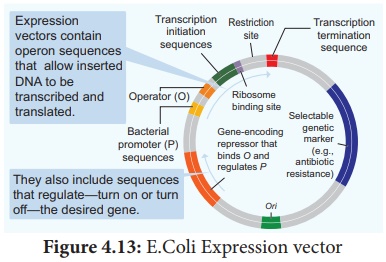
Expression vectors
Vectors which are suitable for expressing foreign
proteins are called expression vectors. This vector consists of signals
necessary for transcription and translation of proteins in the host. This helps
the host to produce foreign protein in large amounts. Example: pUC 19.
More
vectors to know
Cosmid
Cosmids are plasmids containing the ‘cos’ - Cohesive Terminus, the sequence having cohesive ends. They are hybrid vectors derived from plasmids having a fragment of lambda phage DNA with its Cos site and a bacterial plasmid.
Bacteriophage Vectors
Bacteriophages
are viruses that infect bacteria. The most commonly used E. coli phages are l
phage (Lambda phage) and M13 phage. Phage vectors are more efficient than
plasmids - DNA upto 25 Kb can be inserted into phage vector.
Lambda genome: Lambda phage is a temperate bacteriophage that infects Escherichia coli. The genome of lambda-Phage is 48502 bp long, i.e. 49Kb and has 50 genes.
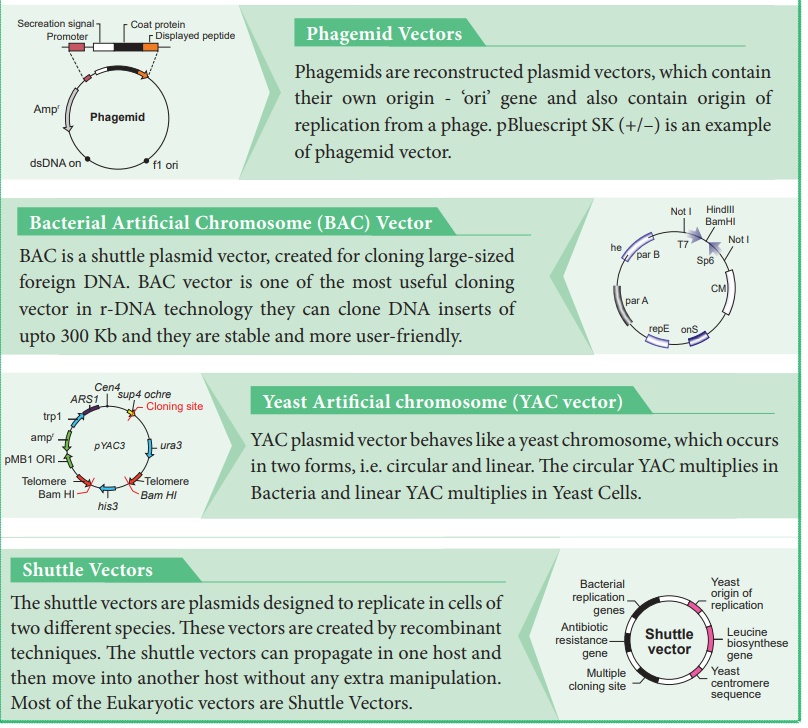
Phagemid Vectors
Phagemids are reconstructed plasmid vectors, which contain their own origin - ‘ori’ gene and also contain origin of replication from a phage. pBluescript SK (+/–) is an example of phagemid vector.
Bacterial Artificial Chromosome (BAC) Vector
BAC is a shuttle plasmid vector, created for cloning large-sized foreign DNA. BAC vector is one of the most useful cloning vector in r-DNA technology they can clone DNA inserts of upto 300 Kb and they are stable and more user-friendly.
Yeast Artificial chromosome (YAC vector)
YAC plasmid vector behaves like a yeast chromosome, which occurs in two forms, i.e. circular and linear. The circular YAC multiplies in Bacteria and linear YAC multiplies in Yeast Cells.
Shuttle Vectors
The
shuttle vectors are plasmids designed to replicate in cells of two different
species. These vectors are created by recombinant techniques. The shuttle
vectors can propagate in one host and then move into another host without any
extra manipulation. Most of the Eukaryotic vectors are Shuttle Vectors.
5. Competent Host (For Transformation with Recombinant DNA)
The propagation of the recombinant DNA molecules must occur inside
a living system or host. Many types of host cells are available for gene
cloning which includes E.coli, yeast, animal or plant cells. The type of host
cell depends upon the cloning experiment. E.coli is the most widely used
organism as its genetic make-up has been extensively studied, it is easy to
handle and grow, can accept a range of vectors and has also been studied for
safety. One more important feature of E.coli to be preferred as a host cell is
that under optimal growing conditions the cells divide every 20 minutes.
Since the DNA is a hydrophilic molecule,it cannot pass through
cell membranes, In order to force bacteria to take up the plasmid, the
bacterial cells must first be made competent to take up DNA. This is done by
treating them with a specific concentration of a divalent cation such as
calcium. Recombinant DNA can then be forced into such cells by incubating the
cells with recombinant DNA on ice, followed by placing them briefly at 420C
(heatshock) and then putting them back on ice. This enables bacteria to take up
the Recombinant DNA.
For the expression of eukaryotic proteins, eukaryotic cells are
preferred because to produce a functionally active protein it should fold
properly and post translational modifications should also occur, which is not
possible by prokaryotic cell (E.coli).
Related Topics