Biotechnology - Screening for Recombinants | 12th Botany : Chapter 4 : Principles and Processes of Biotechnology
Chapter: 12th Botany : Chapter 4 : Principles and Processes of Biotechnology
Screening for Recombinants
Screening for Recombinants
After the introduction of r-DNA into a suitable host cell,
it is essential to identify those cells which have received the r-DNA molecule.
This process is called screening. The vector or foreign DNA present in
recombinant cells expresses the characters, while the non-recombinants do not
express the characters or traits. For this some of the methods are used and one
such method is Blue-White Selection method.
1. Insertional Inactivation - Blue-White Colony Selection Method
It is a powerful method used for screening of recombinant plasmid.
In this method, a reporter gene lacZ is inserted in the vector. The lacZ
encodes the enzyme β-galactosidase and contains several recognition sites for
restriction enzyme.
β-galactosidase breaks a synthetic substrates called X-gal (5-bromo-4-chloro-indolyl-β -D-galacto-pyranoside) into an insoluble blue coloured product. If a foreign gene is inserted into lacZ, this gene will be inactivated. Therefore, no- blue colour will develop (white) because β-galactosidase is not synthesized due to inactivation of lacZ.
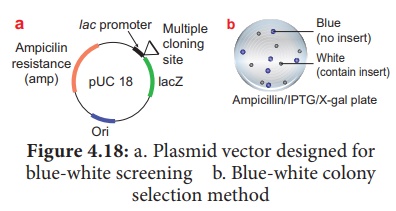
Therefore, the host cell containing r-DNA form white coloured colonies on the medium contain X-gal, whereas the other cells containing
non-recombinant DNA will develop the blue coloured colonies. On the basis of
colony colour, the recombinants can be selected.
2. Antibiotic resistant markers
An antibiotic resistance marker is a gene that produces a protein
that provides cells with resistance to an antibiotic. Bacteria with transformed
DNA can be identified by growing on a medium containing an antibiotic.
Recombinants will grow on these medium as they contain genes encoding
resistance to antibiotics such as ampicillin, chloro amphenicol, tetracycline
or kanamycin, etc., while others may not be able to grow in these media, hence
it is considered useful selectable marker.
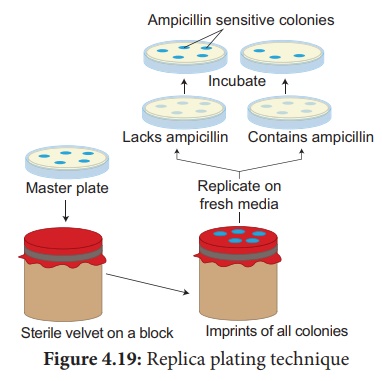
3. Replica plating technique
A technique in which the pattern of colonies growing on a culture
plate is copied. A sterile filter plate is pressed against the culture plate
and then lifted. Then the filter is pressed against a second sterile culture
plate. This results in the new plate being infected with cell in the same
relative positions as the colonies in the original plate. Usually, the medium
used in the second plate will differ from that used in the first. It may
include an antibiotic or without a growth factor. In this way, transformed
cells can be selected.
4. Molecular Techniques - Isolation of Genetic Material and Gel Electrophoresis
Electrophoresis is a separating technique used to separate
different biomolecules with positive and negative charges.
Principle
By applying electricity (DC) the molecules migrate according to
the type of charges they have. The electrical charges on different molecules
are variable.

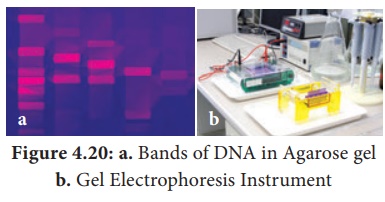
Agarose GEL Electrophoresis
It is used mainly for the purification of specific DNA fragments.
Agarose is convenient for separating DNA fragments ranging in size from a few
hundred to about 20000 base pairs. Polyacrylamide is preferred for the
purification of smaller DNA fragments. The gel is complex network of polymeric
molecules. DNA molecule is negatively charged molecule under an electric field
DNA molecule migrates through the gel. The electrophoresis is frequently
performed with marker DNA fragments of known size which allow accurate size
determination of an unknown DNA molecule by interpolation. The advantages of
agarose gel electrophoresis are that the DNA bands can be readily detected at
high sensitivity. The bands of DNA in the gel are stained with the dye Ethidium
Bromide and DNA can be detected as visible fluorescence illuminated in UV
light will give orange fluorescence, which can be photographed.
Agricultural diagnostics refers to a variety of tests that are
used for detection of pathogens in plant tissues. Two of the most efficient
methods are
1. ELISA (Enzyme Linked Immumo Sorbent Assay)
Elisa is a diagnostic tool for identification of pathogen
species by using antibodies and diagnostic agents. Use of ELISA in plant
pathology especially for weeding out virus infected plants from large scale
planting is well known.
2. DNA Probes
DNA Probes, isotopic and non-isotopic (Northern and Southern
blotting) are popular tools for identification of viruses and other pathogens
5. Nucleic Acid Hybridization - Blotting Techniques
Blotting techniques are widely used analytical tools for the
specific identification of desired DNA or RNA fragments from larger number of
molecules. Blotting refers to the process of immobilization of sample nucleic
acids or solid support (nitrocellulose or nylon membranes.) The blotted nucleic
acids are then used as target in the hybridization experiments for their
specific detection.
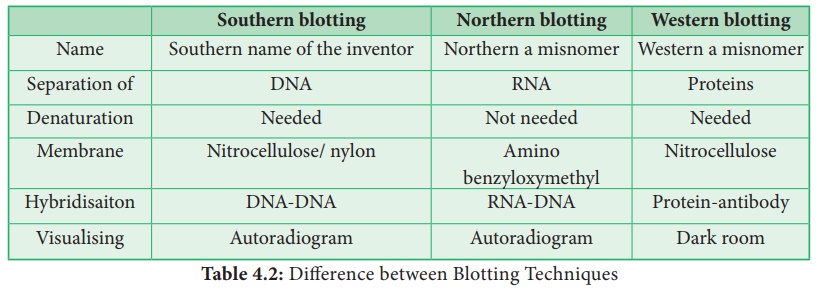
Types of Blotting Techniques
Southern Blotting: The transfer of DNA from agarose gels
to nitrocellulose membrane.
Northern Blotting: The transfer of RNA to nitrocellulose
membrane.
Western Blotting: Electrophoretic transfer of Proteins to
nitrocellulose membrane.
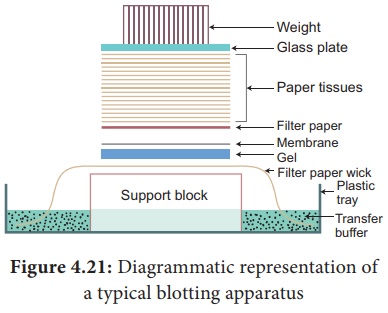
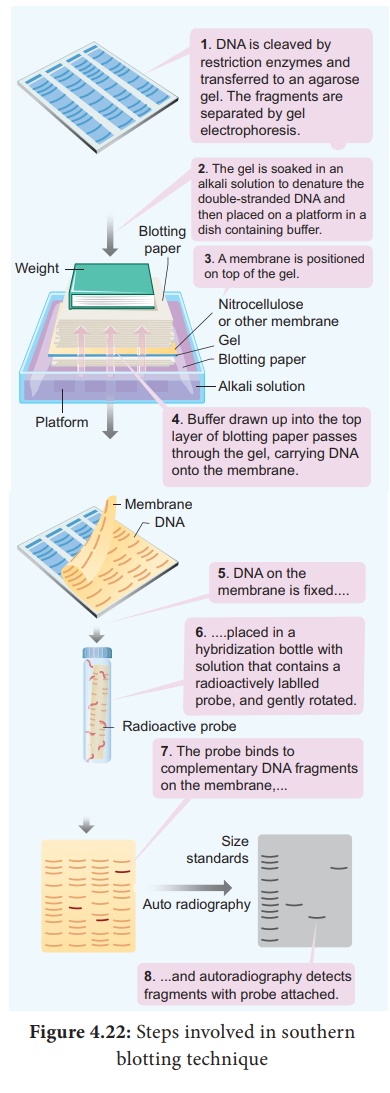
Southern Blotting Techniques - DNA
The transfer of denatured DNA from Agarose gel to Nitrocellulose
Blotting or Filter Paper technique was introduced by Southern in 1975 and this
technique is called Southern Blotting Technique.
Steps
The transfer of DNA from agarose gel to nitrocellulose filter paper
is achieved by Capillary Action.
A buffer Sodium Saline Citrate (SSC) is used, in which DNA is
highly soluble, it can be drawn up through the gel into the Nitrocellulose
membrane.
By this process ss-DNA becomes ‘Trapped’ in the membrane
matrix.
This DNA is hybridized with a nucleic acid and can be detected by
autoradiography.
Autoradiography - A technique that captures the image formed in a
photographic emulsion due to emission of light or radioactivity from a labelled
component placed together with unexposed film.
Northern Blot
It was found that RNA is not binding to cellulose nitrate.
Therefore, Alwin et al. (1979) devised a procedure in which RNA bands
are transferred from the agarose gel into nitrocellulose filter paper. This
transfer of RNA from gel to special filter paper is called Northern Blot
hybridization. The filter paper used for Northern blot is Amino Benzyloxymethyl
Paper which can be prepared from Whatman 540 paper.
Western Blot
Refers to the electrophoretic transfer of proteins to blotting papers. Nitrocellulose filter paper can be used for western blot technique. A particular protein is then identified by probing the blot with a radio-labelled antibody which binds on the specific protein to which the antibody was prepared.
6. Bioassay for Target Gene Effect
Target gene is target DNA, foreign DNA, passenger DNA, exogenous
DNA, gene of interest or insert DNA that is to be either cloned or specifically
mutated. Gene targeting experiments have been targeting the nuclei and this
leads to ‘gene knock-out’. For this purpose, two types of targeting vectors are
used. They are insertion vectors and replacement or transplacement vectors.
1. Insertion vectors are
entirely inserted into targeted locus as the vectors are linearized within the
homology region. Initially, these vectors are circular but during insertion,
become linear. It leads to duplication of sequences adjacent to selectable
markers.
2. The replacement vector
has the homology region and it is co-linear with target. This vector is
linearized prior to transfection outside the homology region and then
consequently a crossing over occurs to replace the endogenous DNA with the
incoming DNA.
Transfection: Introduction of
foreign nucleic acids into cells by non-viral methods.
7. Genome Sequencing and Plant Genome Projects
The whole complement of gene that determine all characteristic of
an organism is called genome. The genome may be nuclear genome, mitochondrial
genome or plastid genome. Genome of many plants contain both functional and
non-expressive DNA proteins. Genome project refer to a project in which the
whole genome of plant is analysed using sequence analysis and sequence homology
with other plants. Such genome projects have so far been undertaken in Chlamydomonas(algae),
Arabidopsis thaliana, rice and maize plants.
Genome content of an organism is expressed in terms of number of
base pairs or in terms of the content of DNA is expressed in c-value.
Genome sequencing: The location of genes
on the entire diploid chromosome of an organism.
8. Evolutionary pattern assessed using DNA.
In recent years the evolutionary relationship between different
plant taxa is assessed using DNA content as well as the similarities and
differences in the DNA sequence (sequence homology). Based on such analysis the
taxa and their relationship are indicated in cladogram. Such cladogram will
show the genetic distance between two taxa. It is also showed antiquity or
modernity of any taxon with respect to one another (See also Unit-2, Chapter-5
of XI Std.)
9. Genome editing and CRISPR - Cas9
Genome editing or gene editing is a group of technologies that has
the ability to change an organism’s DNA. These technologies allow genetic
material to be added, removed, or altered at particular locations in the
genome. Several approaches to genome editing have been developed. A recent one
is known as CRISPR-Cas9, which is short form of Clustered Regularly
Interspaced Short Palindromic Repeats and
CRISPR-associated protein 9. The CRISPR-Cas9 system has generated a lot
of excitement in the scientific community because it is faster, cheaper, more
accurate, and more efficient than other existing genome editing methods.
Rice, was among the first plants to be used to demonstrate the
feasibility of CRISPR-mediated targeted mutagenesis and gene replacement. The
gene editing tool CRISPR can be used to make hybrid rice plants that can clone
their seed. Imtiyaz
Khand and Venkatesan Sundaresan and colleagues
reported in a new study which clearly shows one can re-engineer rice to switch
it from a sexual to an asexual mode.
10. RNA Interference (RNAi)
All characters of organism are the result of expression of
different genes which are regions of nuclear DNA. This expression involves
transcription and translation. Transcription refers to the copying of genetic
information from one strand of the DNA (called sense strand) by RNA. This RNA,
as soon as it formed cannot be straight away sent to the cytoplasm to undertake
the process of translation. It has to be edited and made suitable for
translation which brings about protein synthesis. One of the main items removed
from the RNA strand are the introns. All these changes before translation
normally take place whereby certain regions of DNA are silence. However, there
is an (RNAi) pathway. RNA interference is a biological process in which RNA
molecules inhibit gene expression or translation. This is done by neutralising
targetd mRNA molecules.
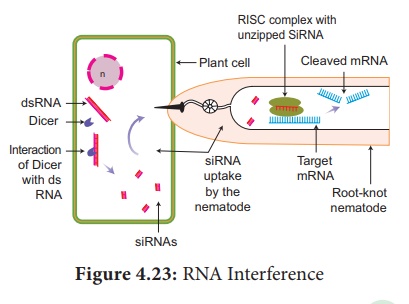
A simplified model for the RNAi pathway is based on two steps, each
involving ribonuclease enzyme. In the first step, the trigger RNA (either dsRNA
or miRNA primary transcript) is processed into a short interfering RNA (siRNA)
by the RNase II enzymes called Dicer and Drosha. In the second step, siRNAs are
loaded into the effector complex RNA-induced silencing complex (RISC). The
siRNA is unwound during RISC assembly and the single- stranded RNA hybridizes
with mRNA target. This RNAi is seen in plant feeding nematodes.
Related Topics