Chapter: Microbiology and Immunology: Bacteriology: Nonsporing Anaerobes
Laboratory Diagnosis of Escherichia coli Infections
Laboratory Diagnosis
Laboratory diagnosis of E. coli infections is based on
a) Isolation of E. coli by culture.
b) Demonstration of toxins of diarrheagenic E. coli.
◗ Specimens
Urine is the specimen of choice for diagnosis of UTI caused by uropathogenic E. coli. Clean-voided midstream samples of urine are usually employed for culture. Catheterized urine and urine collected by suprapubic aspiration are also used in certain situ-ations. Urine is a good medium for the growth of coliforms and other urinary pathogens, hence should be sent immediately to microbiology laboratory for processing the specimen. If delay of more than 1–2 hours is unavoidable, the specimen should be refrigerated. Other specimens include feces or rectal swabs for gastroenteritis, blood for septicemia, cerebrospinal fluid (CSF) for meningitis, sputum for pneumonia, or other body fluids, such as pus from wound, biliary and peritoneal abscesses caused by E. coli.
◗ Culture
Definitive diagnosis is based on the isolation of E. coli from var-ious clinical specimens by culture. Urine culture is a very useful procedure for diagnosis of UTI. Stool culture is widely used to isolate diarrheagenic E. coli. Culture of blood, CSF, and other specimens is also carried out depending on the clinical diseases caused by E. coli, as mentioned earlier.
Urine culture: Semiquantitative culture of urine forE. coliandother Gram-negative bacteria is the method most commonly used in a microbiology laboratory for identification of the bac-teria from urine. In this method, a predetermined quantity of urine is inoculated on MacConkey and blood agar with a stan-dardized inoculating loop. The loop is calibrated to deliver 0.05 mL of urine; hence 200 loopfuls of urine will deliver 1 mL of urine. The number of colonies that are obtained after overnight incubation of inoculated plates multiplied with 200 will be the approximate number of bacteria per milliliter of urine. For example, if the number of colonies on a MacConkey agar after overnight incubation is 500, the viable bacterial count/mL of urine will be (500 200) 100,000 or 105. This forms the basis of significant bacteriuria suggested by Kass.
After incubation overnight at 37°C, pink, flat colonies of E. coli on the MacConkey agar and beta-hemolytic colonies onblood agar are identified by various biochemical tests.
Significant bacteriuria concept suggested by Kass is basedon the fact that a colony count exceeding 100,000 (105) bacteria/mL of urine denotes significant bacteriuria and is suggestive of active UTI. Counts of 10,000 bacteria or less per milliliter are of no significance and are due to contami-nation of urine during voiding. Bacterial counts between 10,000 (104) and 100,000 (105) are infrequent when the sample is collected properly and processed promptly. Such results are considered equivocal and the culture is repeated.
Interpretation of bacteriuria, however, requires caution and should always be with reference to clinical condition of the patient. Because UTI is a common problem and culture facilities are not available everywhere, several simple meth-ods have been introduced for the presumptive diagnosis of significant bacteriuria.
After incubation overnight at 37°C, pink, flat colonies of E. coli on the MacConkey agar and beta-hemolytic colo-nies on blood agar are identified by various biochemical tests.
Other specimens for culture: CSF culture positive forE. coliestablishes the diagnosis of E. coli meningitis. Isolation of the organism from blood, pus, and other specimens is definitive for diagnosis of infections caused by E. coli.
◗ Demonstration of toxins of diarrheagenic E. coli
Laboratory diagnosis of diarrhea caused by diarrheagenic E. coli can be made by demonstration of the bacilli in feces by culture. The feces is collected from the patient in a sterile container and sent immediately to the laboratory. The fecal samples are inoculated directly on MacConkey and blood agar media. The plates are incubated at 37°C overnight and looked for the char-acteristic lactose-fermenting colonies on MacConkey and beta-hemolytic colonies on blood agar as described earlier.
Since E. coli is present as commensals in the intestine—hence is detected even in normal stool—it is essential to perform various diagnostic tests in order to consider it as diarrhea-genic pathogenic E. coli strain. These strains are identified by (a) serotyping, (b) animal inoculation, (c) cytopathic effects in cell cultures, or (d) molecular methods.
Identification of EPEC: Specific serogroups ofE. coli(O26,O55, O86, O111, O114, O119, O125, O126, O12, O128, and O142) are commonly associated with outbreaks of EPEC. Therefore, E. coli colonies isolated from feces on MacConkey agar are identified by agglutination tests with specific poly-valent and monovalent antisera. In this method, a saline suspension of E. coli colonies is made on the slide and mixed with a drop of specific polyvalent and monovalent antisera against EPEC serogroups. At least 10 colonies per plate should be tested. If isolated colonies are negative, the con-fluent growth is emulsified and tested. In a positive test, if E. colicolonies show agglutination with a specific serogroup(for example, O111) then the isolate is identified as E. coli of that serogroup (O111).
Identification of ETEC: Diagnosis of ETEC diarrhea dependson the demonstration of enterotoxin in E. coli isolates from stool, because toxin production is not associated with specific serogroups of E. coli. A strain of ETEC may produce either LT or ST, or both. Many tests are available for demonstration of the LT or ST produced by ETEC. These tests, therefore, are used for detection and identification of E. coli isolates from stool as ETEC (Table 31-7).
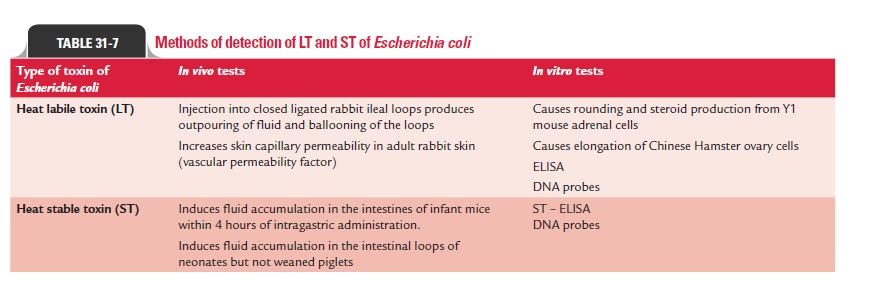
1. The presence of LT in isolates of E. coli can be demonstrated by:
· Showing fluid accumulation in rabbit ileal loop method.
· Showing vascular permeability factor of the toxin in adult rabbit skin method.
· Tissue culture tests (rounding of Y1 mouse adrenal cells and elongation of Chinese hamster ovary cells [CHO] cells due to intracellular increase of cAMP concentration).
· Serological tests (agar gel diffusion, reverse passive hem-agglutination, and enzyme-linked immunosorbent assay).
· Genetic probes. These are available for detection of LT in E. coli isolates from stool cultures, or directly in feces.
2. The presence of ST in isolates of E. coli can be demonstrated by:
· Infant mouse test—it is still widely employed for detec-tion of ST in isolates of E. coli.
· Genetic probes—for detection of ST in E. coli isolates from stool cultures, or directly in feces.
Identification of EIEC: Many of the EIEC strains are atypicalE. coli strains. They are nonmotile and do not ferment lactose,or ferment it late with production of acid, but without produc-ing any gas. They also do not decarboxylate lysine. EIEC are identified by:
Sereny test: The test is carried out in guinea pigs by instillationof isolated EIEC into the conjunctival sac of guinea pigs. The animal is examined after 72 hours for mucopurulent conjunc-tivitis and severe keratitis.
Cell culture test: The toxin can be demonstrated in HeLa orHEP-2 cells. The bacterial suspension is added to a monolayer of the cells. The cells are then examined for the presence of intracellular E. coli because EIEC, if present, penetrate these cells and replicate inside the cells.
VMA enzyme-linked immunosorbent assay: This is a serologicaltest used to detect the plasmid, which codes for outer mem-brane antigens called the VMA, in stool isolates of EIEC.
Identification of EHEC: E. coliO157:H7 is the most commonserotype associated with the clinical disease caused by EHEC strains. The strain typically does not ferment sorbitol; hence sorbitol MacConkey medium is frequently used for isolation of the strain from stool by culture. Identification of EHEC strains is by:
· Demonstrating cytotoxic effects of EHEC on Vero or HeLa cells.
· Using DNA probes for VT1 and VT2 genes in EHEC directly in feces or in culture isolates.
Identification of other strains: EAEC strains are identified byagglutination tests with specific antisera. Most of them are not typed by O antisera, but by specific H antisera.
Related Topics