Chapter: Genetics and Molecular Biology: Genetics
Genetics Mapping by Recombination Frequencies
Mapping by Recombination Frequencies
Two or more mutations on any DNA molecule that
engages in recom-bination can be approximately ordered along DNA molecules by
meas-urement of the frequencies of recombination. Let us examine why this is
so. Assume that the probability of a recombination or genetic cross - over
between two points on two almost homologous DNA molecules is a function only of
the distance between the points. Up to a limit, the greater the separation, the
greater the probability of crossover. This seems reasonable because increasing
the separation increases the num-ber of potential crossover sites.
Recombination frequencies should be additive if the frequency is linearly
proportional to marker separation. Thus, if Rf(X,Y) is the recombination
frequency between markers X and
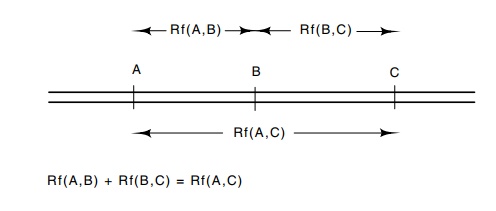
Figure
8.8 Additivity relations necessary
when recombination frequency isnot linearly proportional to genetic distance.
Y, and if strict additivity holds, Rf(A,C) must
equal Rf(A,B) + Rf(B,C) (Fig. 8.8).
Unfortunately, the assumption of a linear relation
between recombi-nation frequency and distance is not good over extremely short
or long distances. At short distances the specific nucleotides involved can
gen-erate profound effects on the recombination frequencies, and additivity
fails as a result. These short-distance anomalies are often called marker
effects. At large distances more than one crossover is likely in the distance
separating two markers. If an even number of crossovers occurs between the
markers, no net recombination is observed between the markers. Therefore, as
the distance between markers becomes large and multiple crossovers occur, about
as many partners experience an even number of crossovers as experience an odd
number. As only those with odd numbers of crossovers generate recombinants,
recombination frequencies between two markers rise to 50% as an upper limit.
Given the vagaries of measuring distances by
recombination frequen-cies, what can be done? Often the major question is
merely one of marker order. The actual distances separating the markers are not
of great importance. Three-factor genetic crosses are a partial solution. They
permit ordering of two genetic markers, B
and C, with respect to a third, A, that is known to lie on the outside.
That is, the experiment is to determine whether the order is A-B-C or A-C-B.
Here the crosses will be described as they are
performed in prokary-otes in which only one of the two participating DNA
molecules is a complete chromosome, but the basic principles apply for the
situation of recombination between two complete chromosomes. Mating
experi-ments permit the introduction of a portion of the chromosomal DNA from a
donor bacterial cell into a recipient cell in a process called bacterial
conjugation. As a result of the inviability of such a linear DNA fragment, if a
crossover occurs between the chromosome of the recipi-ent and the incoming
chromosome fragment, a second crossover be-tween the two DNAs must also occur
for the recipient chromosome not to be left open by the single crossover event
(Fig. 8.9). In general, any even number of total crossovers between the
incoming DNA and the recipient chromosome will yield viable recombinants.
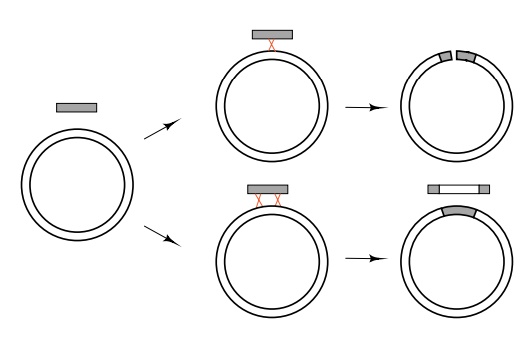
Figure
8.9 In order that a circular
chromosome not be opened by geneticrecombination, two crossovers between it and
a fragment of linear DNA are necessary.
The
genetic markers involved in the genetic cross, A, B, and C, can be different genes or different
alleles within the same gene. In principle it does not matter, but in practice
it could be difficult to ascertain the different varieties of recombinants if
the markers all lie within the same gene.
To
determine the gene order, compare the fraction of B+C+
recombi-nants among all the A+
recombinants in the two crosses (Fig. 8.10), first between an A-B-C+
chromosome and an A+B+C- DNA fragment, and then between an A-B+C-
chromosome and an A+B-C+ fragment. If the gene order is ABC, then generation of B+C+ recombinants from the
first cross requires only two crossover events, whereas the second cross
requires crossovers in the same two intervals as the first plus an additional
two crossovers. Therefore the fraction of the B+C+
recombinants will be much higher from the first cross. Alternatively, if the
gene order is ACB,
Figure
8.10 Three-factor crosses. The first
requires only two crossovers toproduce A+B+C+ progeny whereas the second requires four.
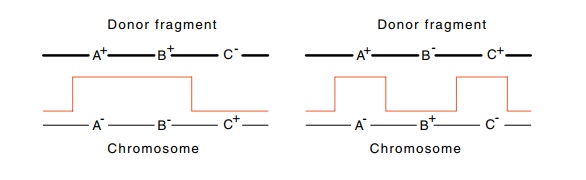
Figure 8.11 Recombination be-tween a chromosome containing a point mutation and a DNA frag-ment containing a deletion. Crossovers can occur only be-tween homologous segments, that is, outside a deleted area.a greater fraction of B+C+

Related Topics