Chapter: 11th Microbiology : Chapter 13 : Immunology
Structure of an Immunoglobulin Molecule
Structure of an Immunoglobulin
Molecule
1. Basic unit
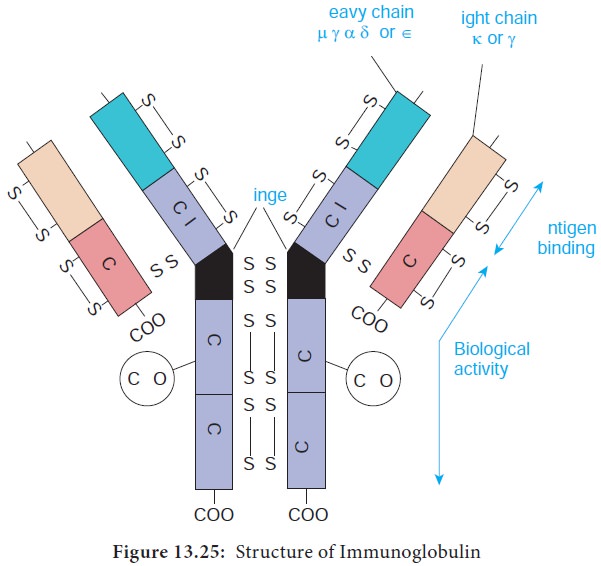
The basic structural unit (monomer) of an immunoglobulin
molecule consists of four polypeptide chains linked covalently by disulfide
bonds (Figure 13.25). The four-chain structure is composed of two identical light
(L) and two identical heavy (H) polypeptide chains. Every immunoglobulin can be
represented by the general formula (H2L2)n.
a) Light chains
Light Chains have a molecular weight of approximately 25000 Da and are composed of about 220 amino acids. Light chains are common to all immunoglobulin classes and are of two types – kappa (κ) or lambda(λ) - based on their structural differences. A given immunoglobulin molecule may contain either identical κ or λ chains but never both.
b) Heavy chains
Heavy chains have a molecular weight of approximately twice that
of light chains (57000-70000 Da) and twice the number of amino acids (about
440). Five antigenically distinct isotypes of heavy chains are recognized-gamma
(γ), alpha (α), mu (μ), delta (δ) and epsilon (ε) – based on structural
differences in the carboxy terminal portion of heavy chains. The heavy chains
isotypes form the basis of five classes of immunoglobulin molecules – IgG
(contains γ chain), IgA (contains α chain), IgM (contains μ chain), IgD
(contains δ chain) and IgE (contains ε chain). Five heavy chain classes of
immunoglobulin can be easily remembered as GAMDE. Heavy chain classes are again
subdivided into subclasses of molecules.
1.
Four known subclasses of the γ chain exist – γ1, γ2, γ3 and γ4 -
which yield IgG1, IgG2, IgG3 and IgG4.
2.
Two subclasses of the α chain are known – α1 and α2 - which
yield IgA1 and IgA2.
3.
Two subclasses of the μ chain are known – μ1 and μ2 - which
yield IgM1 and IgM2.
4.
No subclasses of the δ and ε (IgD and IgE) are known.
2. Disulfide bonds
Disulfide bonds hold together the four polypeptide chains in
normal immunoglobulin molecules and are of two types namely interchain bonds and intrachain bonds.
a. Inter chain bonds occur between heavy chains (H-H), heavy and light chains (H-L) and light
chains (L-L). H-H bonds occur primarily in the hinge region and can vary in
number from 1-15 depending on the class and subclass of the immunoglobulin
molecules.
b. Intra chain bonds are stronger than interchain bonds and occur within the individual chain type,
with the number of bonds varying depending on the type (light chains have two,
human γ, α and δ heavy chains have four and human μ and ε heavy chains have
five). The distribution of intrachain disulfide bonds forms the basis for
division of each immunoglobulin into domains.
3. Regions
Each heavy and light chain consists of two segments, the variable region and the constant region. The variable (V) region shows a wide variation in amino acid sequence in the
amino terminal portion of the molecule. The areas of high variability in the
variable region of heavy (VH) and light (VL) chains are called hypervariable regions or complementarity determining
regions (CDRs). Hypervariable regions are most intimately involved in
formation of the antigen binding site.
4. Domains
Each immunoglobulin chain consists of a series of globular
regions enclosed by disulphide bonds. Each heavy chain consists of four or five
domains - one in the variable region (VH ) and three or four in the constant
region (CH1, CH2, CH3, and CH4). Each light chain consists of two domains – one
in the variable region (VL) and one in the constant region (CL).
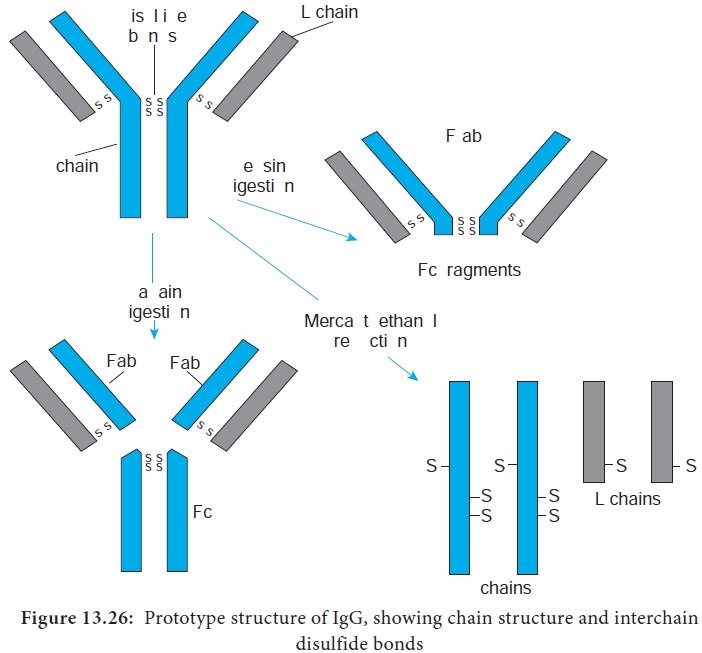
5. Fragments.
Proteolytic (peptide bond -splitting) enzymes such as papain and pepsin are used to degrade
immunoglobulin molecules into definable fragments to facilitate study of their
structure.
i. Treatment of the monomeric basic unit with the enzyme papain
splits it into two Fab fragments (Fragment-antigen binding) and one Fc fragment.
These Fab fragments can bind but cannot precipitate the antigen; therefore,
they are monovalent, possessing only one combining site each.
ii. Treatment of the immunoglobulin molecule with pepsin results
in digestion of most of the Fc fragment, leaving one large fragment that
consists of two Fab fragments joined by covalent bonds, termed the F(ab’)2 fragment. The F(ab’)2 fragments has two antigen
combining sites. Therefore it is bivalent, possessing the ability to bind and
precipitate an antigen (Figure 13.26).
6. Hinge region
Hinge region is the portion of
heavy chain between the CH1 and
CH2 domains. It is highly flexible and allows for movement of the Fab arms in
relation to each other. The S values (sedimentation coefficient that is expressed in Svedberg units(s)) of immunoglobulins range from 7S- 19S.
Related Topics