Molecular Genetics - Transcription | 12th Zoology : Chapter 5 : Molecular Genetics
Chapter: 12th Zoology : Chapter 5 : Molecular Genetics
Transcription
Transcription
Francis Crick proposed
the Central dogma in molecular biology which states that genetic
information flows as follows:
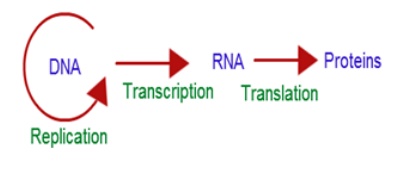
The process of copying
genetic information from one strand of DNA into RNA is termed transcription.
This process takes place in presence of DNA dependent RNA polymerase.
In some retroviruses
that contain RNA as the genetic material (e.g, HIV), the flow of information is
reversed. RNA synthesizes DNA by reverse transcription, then transcribed into
mRNA by transcription and then into proteins by translation. ![]()
![]()
For a cell to operate,
its genes must be expressed. This means that the gene products, whether
proteins or RNA molecules must be made. The RNA that carries genetic
information encoding a protein from genes into the cell is known as messenger
RNA (mRNA). For a gene to be transcribed, the DNA which is a double helix must
be pulled apart temporarily, and RNA is synthesized by RNA polymerase. This
enzyme binds to DNA at the start of a gene and opens the double helix. Finally,
RNA molecule is synthesized. The nucleotide sequence in the RNA is
complementary to the DNA template strand from which it is synthesized.
Both the strands of DNA
are not copied during transcription for two reasons. 1. If both the strands act
as a template, they would code for RNA with different sequences. This in turn
would code for proteins with different amino acid sequences. This would result
in one segment of DNA coding for two different proteins, hence complicate the
genetic information transfer machinery. 2. If two RNA molecules were produced
simultaneously, double stranded RNA complementary to each other would be
formed. This would prevent RNA from being translated into proteins.
1. Transcription unit and gene
A transcriptional unit
in DNA is defined by three regions, a promoter, the structural gene
and a terminator. The promoter is located towards the 5' end. It is a
DNA sequence that provides binding site for RNA polymerase. The presence of
promoter in a transcription unit, defines the template and coding strands. The
terminator region located towards the 3' end of the coding strand contains a
DNA sequence that causes the RNA polymerase to stop transcribing. In eukaryotes
the promoter has AT rich regions called TATA box (Goldberg
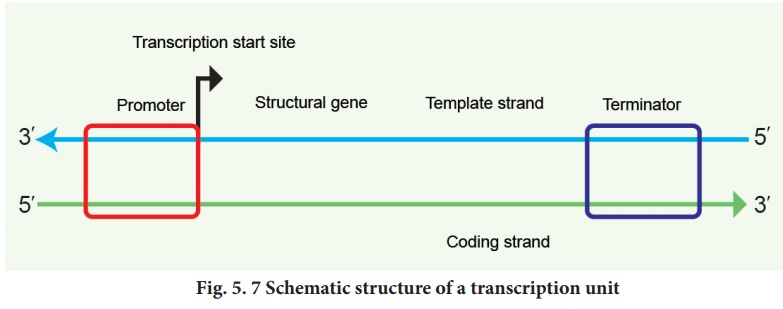
The two strands of the
DNA in the structural gene of a transcription unit have opposite polarity. DNA
dependent RNA polymerase catalyses the polymerization in only one direction,
the strand that has the polarity 3'→5' acts as a template, and is called the template
strand. The other strand which has the polarity 5'→3' has a sequence same
as RNA (except thymine instead of uracil) and is displaced during transcription.
This strand is called coding strand (Fig. 5.7).
The structural gene may
be monocistronic (eukaryotes) or polycistronic (prokaryotes). In
eukaryotes, each mRNA carries only a single gene and encodes information for
only a single protein and is called monocistronic mRNA. In prokaryotes,
clusters of related genes, known as operon, often found next to each other on
the chromosome are transcribed together to give a single mRNA and hence are
polycistronic.
Before starting
transcription, RNA polymerase binds to the promoter, a recognition sequence in
front of the gene. Bacterial (prokaryotic) RNA polymerase consists of two major
components, the core enzyme and the sigma subunit. The core enzyme (β1,
β, and α) is responsible for RNA
synthesis whereas a sigma subunit is responsible for recognition of the
promoter. Promoter sequences vary in different organisms.
RNA polymerase opens up
the DNA to form the transcription bubble. The core enzyme moves ahead,
manufacturing RNA leaving the sigma subunit behind at the promoter region. The
end of a gene is marked by a terminator sequence that forms a hair pin
structure in the RNA. The sub-class of terminators require a recognition
protein, known as rho (ρ), to function.
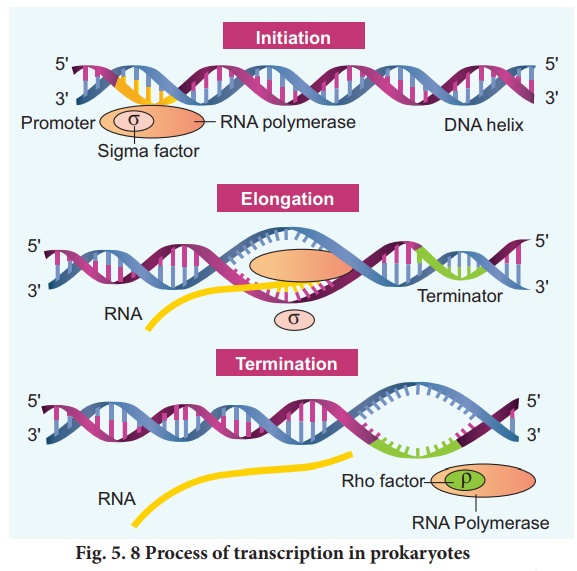
2. Process of transcription
In prokaryotes, there
are three major types of RNAs: mRNA, tRNA, and rRNA. All three RNAs are needed
to synthesize a protein in a cell. The mRNA provides the template, tRNA brings
amino acids and reads the genetic code, and rRNAs play structural and catalytic
role during translation. There is a single DNA-dependent RNA polymerase that
catalyses transcription of all types of RNA. It binds to the promoter and initiates
transcription (Initiation). The polymerases binding sites are called promoters.
It uses nucleoside triphosphate as substrate and polymerases in a template
depended fashion following the rule of complementarity. After the initiation of
transcription, the polymerase continues to elongate the RNA, adding one
nucleotide after another to the growing RNA chain. Only a short stretch of RNA
remains bound to the enzyme, when the polymerase reaches a terminator at the
end of a gene, the nascent RNA falls off, so also the RNA polymerase.
The question is, how the
RNA polymerases are able to catalyse the three steps initiation, elongation and
termination? The RNA
The RNA polymerase associates
transiently with initiation factor sigma (σ) and termination factor rho (r) to
initiate and terminate the transcription, respectively.Association of RNA with
these factors instructs the RNA polymerase either to initiate or terminate the
process of transcription (Fig. 5.8).In bacteria,since the mRNA does not
require any processing to become active and also since transcription
and translation take place simultaneously in the same compartment (since there
is no separation of cytosol and nucleus in bacteria), many times the
translation can begin muc before the mRNA is fully transcribed. This is because
the genetic material is not separated from other cell organelles by a nuclear
membrane consequently; transcription and translation can be coupled in
bacteria.
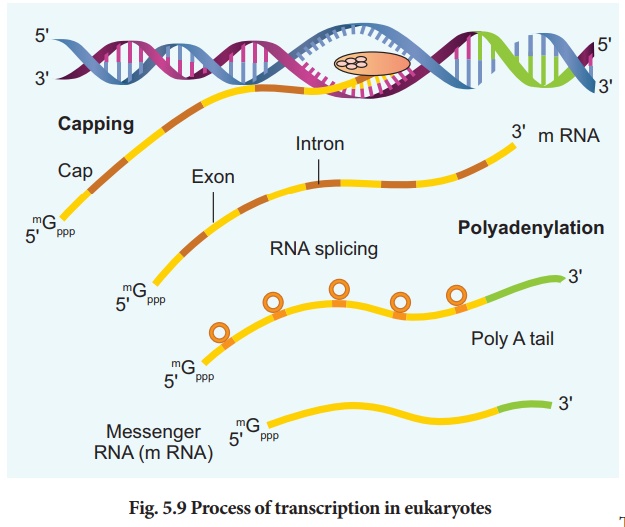
In Eukaryotes, there are
at least three RNA polymerases in the nucleus (in addition to RNA polymerase
found in the organelles). There is a clear division of labour. The RNA
polymerase I transcribes rRNAs (28S, 18S and 5.8S), whereas the RNA polymerase
III is responsible for transcription of tRNA, 5srRNA and snRNA. The RNA
polymerase II transcribes precursor of mRNA, the hnRNA (heterogenous nuclear
RNA). In eukaryotes, the monocistronic structural genes have interrupted coding
sequences known as exons (expressed sequences) and non-coding sequences called
introns (intervening sequences). The introns are removed by a process called
splicing. hnRNA undergoes additional processing called as capping and tailing.
In capping an unusual nucleotide, methyl guanosine triphosphate is added at the
5' end, whereas adenylate residues (200-300) (Poly A) are added at the 3' end
in tailing (Fig. 5.9). Thereafter, this processed hnRNA, now called mRNA is
transported out of the nucleus for translation.
The split gene feature
of eukaryotic genes is almost
entirely absent in
prokaryotes. Originally each exon may have coded for a single
polypeptide chain with a specific function. Since exon arrangement and intron
removal are flexible, the exon coding for these polypeptide subunits act as
domains combining in various ways to form new genes. Single genes can produce
different functional proteins by arranging their exons in several
different ways through
alternate splicing
Introns would have arosen before or after the evolution
of eukaryotic gene. If introns arose late how did they enter eukaryotic gene?
Introns are mobile DNA sequences that can splice themselves out of, as well as
into, specific ‘target sites’ acting like mobile transposon-like elements (that
mediate transfer of genes between organisms– Horizontal Gene Transfer - HGT).
HGT occurs between lineages of prokaryotic cells, or from prokaryotic to
eukaryotic cells and between eukaryotic cells. HGT is now hypothesized to have
played a major role in the evolution of life on earth.
Related Topics