Molecular Genetics - Regulation of gene expression | 12th Zoology : Chapter 5 : Molecular Genetics
Chapter: 12th Zoology : Chapter 5 : Molecular Genetics
Regulation of gene expression
Regulation
of gene expression
We have previously
established how DNA is organized into genes, how genes store genetic
information, and how this information is expressed. We now consider the most
fundamental issues in molecular genetics. How is genetic expression regulated?
Evidence in support of the idea that genes can be turned on and off is very
convincing. Regulation of gene expression has been extensively studied in
prokaryotes, especially in E. coli. Gene expression can be controlled or
regulated at transcriptional or post transcriptional or translational level.
Here, we are going to discuss regulation of gene expression at transcriptional
level. Usually, small extracellular or intracellular metabolites trigger
initiation or inhibition of gene expression. The clusters of gene with related
functions are called operons. They usually transcribe single mRNA
molecules.
In E.coli, nearly 260
genes are grouped into 75 different operons.
Structure of the operon: Each operon is a unit of gene expression and
regulation and consists of one or more structural genes and an adjacent
operator gene that controls
transcriptional activity of the structural gene.
i) The
structural gene codes for proteins, rRNA and tRNA required by the cell.
ii) Promoters are
the signal sequences in DNA that initiate RNA synthesis. RNA polymerase binds
to the promoter prior to the initiation of transcription.
iii) The operators are
present between the promoters and structural genes. The repressor protein binds
to the operator region of the operon.
The Lac (Lactose) operon: The metabolism of lactose in E.coli requires
three enzymes – permease,
β-galactosidase (β-gal) and transacetylase. The
enzyme permease is needed for entry of lactose into thecell,
β-galactosidase brings about hydrolysis of lactose to glucose and
galactose, while transacetylase transfers acetyl group from acetyl Co A
to β-galactosidase.
The lac operon consists
of one regulator gene (‘i’ gene refers to inhibitor) promoter sites (p),
and operator site (o). Besides these, it
has three structural genes namely
lac z,y and lac a. The lac ‘z’ gene codes for β-galactosidase, lac ‘y’
gene codes for permease and ‘a’ gene codes for transacetylase.
Jacob and Monod proposed
the classical model of Lac operon to explain gene expression and regulation in
E.coli. In lac operon, a polycistronic structural gene is regulated by a common
promoter and regulatory gene. When the cell is using its normal energy source
as glucose, the ‘i’ gene transcribes a repressor mRNA and after its
translation, a repressor protein is produced. It binds to the operator region
of the operon and prevents translation, as a result, β-galactosidase is not
produced. In the absence of preferred carbon source such as glucose, if lactose
is available as an energy source for the bacteria then lactose enters the cell
as a result of permease enzyme. Lactose acts as an inducer and interacts with
the repressor to inactivate it.
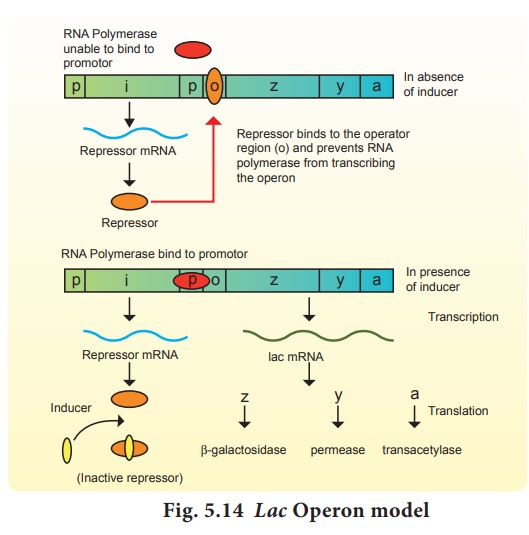
The repressor protein
binds to the operator of the operon and prevents RNA polymerase from
transcribing the operon. In the presence of inducer, such as lactose or
allolactose, the repressor is inactivated by interaction with the inducer. This
allows RNA polymerase to bind to the promotor site and transcribe the operon to
produce lac mRNA which enables formation of all the required enzymes needed for
lactose metabolism (Fig. 5.14).
This regulation of lac operon by the repressor is an example
of negative control of transcription initiation. Lac
Related Topics