Molecular Genetics - Mechanism of Translation | 12th Zoology : Chapter 5 : Molecular Genetics
Chapter: 12th Zoology : Chapter 5 : Molecular Genetics
Mechanism of Translation
Translation
Translation refers to
the process of polymerization of amino acids to form poly peptide chain. The
decoding process is carried out by ribosomes that bind mRNA and charged tRNA
molecules. The mRNA is translated, starting at the 5' end. After binding to
mRNA, the ribosomes move along
it, adding new amino acids to the growing polypeptide
chain each time it reads a codon. Each codon is read by an anticodon on the
corresponding tRNA. Hence the order and sequence of amino acids are defined by
the sequence of bases in the mRNA.
Mechanism of Translation
The cellular factory
responsible for synthesizing protein is the ribosome. The ribosome consists of
structural RNAs and about 80 different proteins. In inactive state, it exists
as two subunits; large subunit and small subunit. When the subunit encounters
an mRNA, the process of translation of the mRNA to protein begins. The
prokaryotic ribosome (70 S) consists of two subunits, the larger subunit (50 S)
and smaller subunit (30 S). The ribosomes of eukaryotes (80 S) are larger,
consisting of 60 S and 40 S sub units. ‘S’ denotes the sedimentation efficient
which is expressed as Svedberg unit (S). The 30 S subunit of bacterial ribosome
contains 16Sr RNA and 50 S subunit contains 5Sr RNA molecules and 23 S RNA and
31 ribosomal proteins. The larger subunit in eukaryotes consist of a 23 S RNA
and 5Sr RNA molecule and 31 ribosomal proteins. The smaller eukaryotic subunit
consist of 18Sr RNA component and about 33 proteins.
One of the alternative ways of dividing up a sequence of bases in DNA or RNA into codons is called reading frame. Any sequence of DNA or RNA, beginning with a start codon and which can be translated into a protein is known as an Open Reading Frame (ORF). A translational unit in mRNA is the sequence of RNA that is flanked by the start codon (AUG) and the stop codon and codes for polypeptides.
mRNA also have some additional
sequences that are not translated and are referred to as Untranslated Regions
(UTR). UTRs are present at both 5' end (before start codon) and at
3' end (after stop codon). The start codon (AUG) begins the coding
sequence and is read by a special tRNA that carries methionine (met). The
initiator tRNA charged with methionine binds to the AUG start codon. In
prokaryotes, N - formyl methionine (f met ) is attached to the
initiator tRNA whereas in eukaryotes unmodified methionine is used. The 5' end
of the mRNA of prokaryotes has a special sequence which precedes the initial
AUG start codon of mRNA. This ribosome binding site is called the Shine –
Dalgarno sequence or S-D sequence. This sequences base -pairs with a
region of the 16Sr RNA of the small ribosomal subunit facilitating initiation.
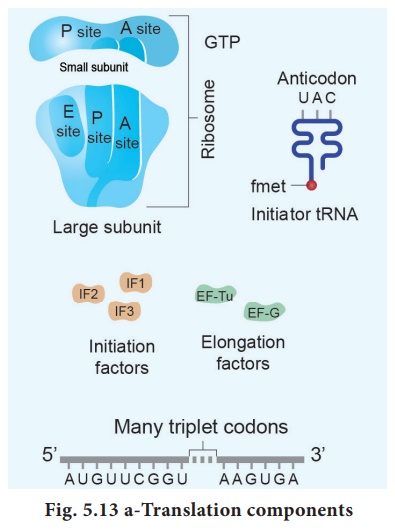
The subunits of the ribosomes (30 S and 50 S) are usually dissociated from each
other when not involved in translation (Fig. 5.13a).
Initiation of translation in E.
coli begins with the formation of an initiation complex,
consisting of the 30S subunits of the ribosome, a messenger RNA and the charged
N-formyl methionine tRNA (fmet – t RNA fmet), three
proteinaceous initiation factors (IF1, IF2, IF3), GTP(Guanine Tri Phosphate)
and Mg 2+.
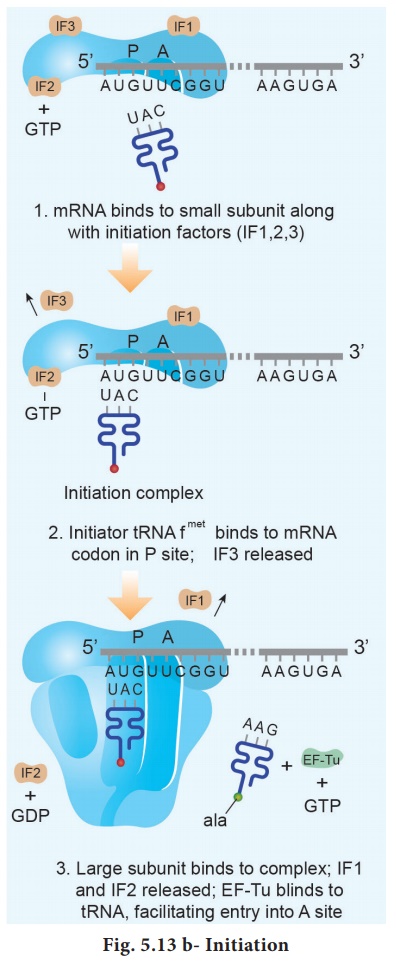
The components that form
the initiation complex interact in a series of steps. IF3 binds to the 30S and
allows the 30S subunit to bind to mRNA. Another initiation protein (IF2) then
enhances the binding of charged formyl methionine tRNA to the small subunit in
response to the AUG triplet. This step ‘sets’ the reading frame so that all
subsequent groups of three ribonucleotides are translated accurately.
The assembly of
ribosomal subunits, mRNA and tRNA represent the initiation complex. Once initiation
complex has been assembled, IF3 is released and allows the
initiation complex to combine with the 50S ribosomal subunit to form the
complete ribosome (70S). In this process a molecule of GTP is hydrolyzed
providing the required energy and the initiation factors (IF2 and IF2 and GDP)
are released (Fig. 5.13 b).
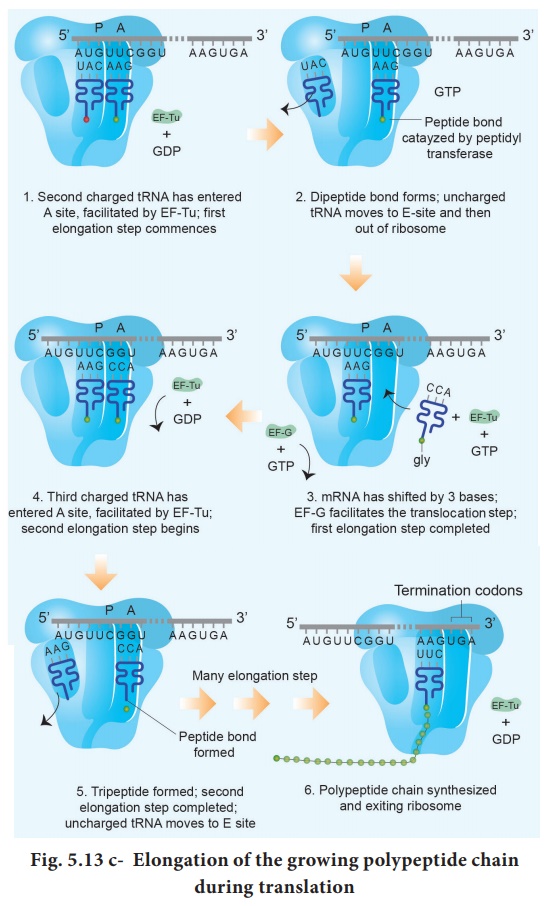
Elongation is the second phase of translation. Once both subunits of the ribosomes are assembled with the mRNA, binding sites for two charged tRNA molecules are formed. The sites in the ribosome are referred to as the aminoacyl site (A site), the peptidyl site (P site) and the exit site (E site). The charged initiator tRNA binds to the P site. The next step in prokaryotic translation is to position the second tRNA at the ‘A’ site of the ribosome to form hydrogen bonds between its anticodon and the second codon on the mRNA (step1). This step requires the correct transfer RNA, another GTP and two proteins called elongation factors (EF-Ts and EF-Tu).
Once the charged tRNA
molecule is positioned at the A site, the enzyme peptidyl transferase catalyses
the formation of peptide bonds that link the two amino acids together (step 2).
At the same time, the covalent bond between the amino acid and tRNA occupying
the P site is hydrolyzed (broken). The product of this reaction is a dipeptide
which is attached to the 3' end of tRNA still residing in the A site. For
elongation to be repeated, the tRNA attached to the P site, which is now
uncharged is released from the large subunit. The uncharged tRNA moves through
the ‘E’ site on the ribosome.
The entire
mRNA-tRNA-aa1-aa2 complex shifts in the direction of the ‘P’ site by a
distance of three nucleotides (step 3). This step requires several elongation
factors (EFs) and the energy derived from hydrolysis of GTP. This results in
the third triplet of mRNA to accept another charged tRNA into the A site (step
4). The sequence of elongation is repeated over and over (step 5 and step 6).
An additional amino acid is added to the growing polypeptide, each time mRNA
advances through the ribosome. Once a polypeptide chain is assembled, it emerges
out from the base of the large subunit (Fig. 5.13 c).
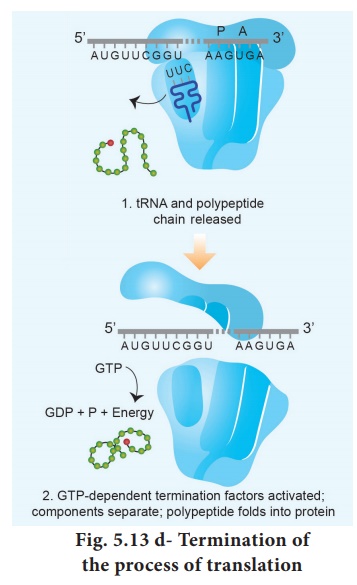
Termination is the third phase of translation. Termination of protein synthesis occurs when one of the three stop codons appears in the ‘A’ site of the
ribosome. The terminal codon signals the action of GTP – dependent release
factor, which cleaves the polypeptide chain from the terminal
tRNA releasing it from the translational complex (step 1). The tRNA is then
released from the ribosome, which then dissociates into its subunits (step 2) (Fig.
5.13 d).
Many antibiotics do
not allow pathogenic bacteria to flourish in animal host because they inhibit
one or the other stage of bacterial protein synthesis. The antibiotic
tetracycline inhibits binding between aminoacyl tRNA and mRNA. Neomycin inhibits
the interaction between tRNA and mRNA. Erythromycin inhibits the translocation
of mRNA along the ribosome. Streptomycin inhibits the initiation of translation
and causes misreading. Chloramphenicol inhibits peptidyl transferase and
formation of peptide bonds.
Related Topics