Chapter: Biotechnology Applying the Genetic Revolution: Protein Engineering
Structural Scaffolds
STRUCTURAL
SCAFFOLDS
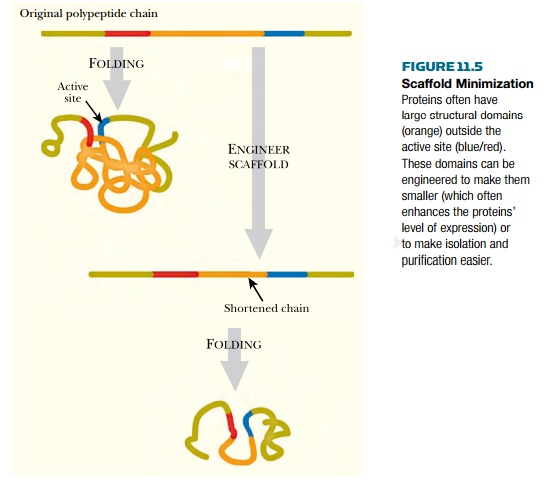
Relatively few of the amino
acid residues in a protein are actually involved in the active site. Most of
the protein provides the 3D platform or scaffold needed to correctly position
the active site residues. Quite often the scaffold is much larger than really
necessary. For example, the β-galactosidase of Escherichia coli (LacZ protein) has
approximately 1000 amino acids, whereas most simple hydrolytic enzymes have
only 200 to 300. Presumably it should be possible to redesign a functional β-galactosidase that is only 25% to 30% the size
of LacZ protein. From an industrial viewpoint, such a smaller protein would
obviously be more efficient.
The concept of using only the
active site region of useful proteins has already been applied to antibody
engineering as discussed. In addition, the technique of phage display has been used to select relatively short
peptides for binding to a variety of target molecules. Once such a binding
domain has been selected, it can be grafted on to another protein. Such techniques
will allow the engineering of smaller proteins whose biosynthesis consumes less
energy and material (Fig. 11.5).
Related Topics