Chapter: Biotechnology Applying the Genetic Revolution: Protein Engineering
Recombining Domains
RECOMBINING
DOMAINS
A related approach to
directed evolution is to deliberately recombine functional domains from
different proteins. An example is the creation of novel restriction enzymes by
linking the cleavage domain from the restriction enzyme FokI with different sequence-specific DNA binding domains. FokI is a type II restriction enzyme with distinct N-terminal and C-terminal
domains that function in DNA recognition and DNA cutting, respectively.
By itself the endonuclease
domain cuts DNA nonspecifically. However, when the nuclease domain is attached
to a DNA-binding domain, this domain determines the sequence specificity of the
hybrid protein. The two domains may be joined via a sequence encoding a linker
peptide such as (GlyGlyGlyGlySer)3 (Fig. 11.8). Cleavage of the DNA
occurs several bases to the side of the recognition sequence, as in the native
FokI restriction enzyme.
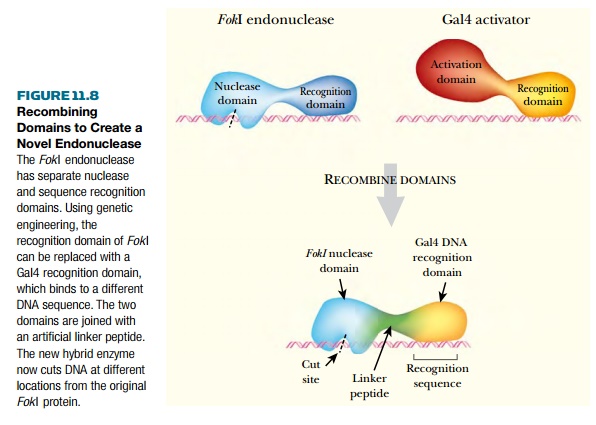
Several different DNA binding domains have been combined with the nuclease domain of FokI. For example, the Gal4 protein of yeast is a transcriptional activator that recognizes a 17 base-pain consensus sequence. Gal4 has two domains, a DNA binding domain and a transcription activating domain. The N-terminal 147 amino acids of Gal4 can be fused to the nuclease domain of FokI, giving a hybrid protein that binds to the Gal4 consensus sequence and cleaves the DNA at that location.
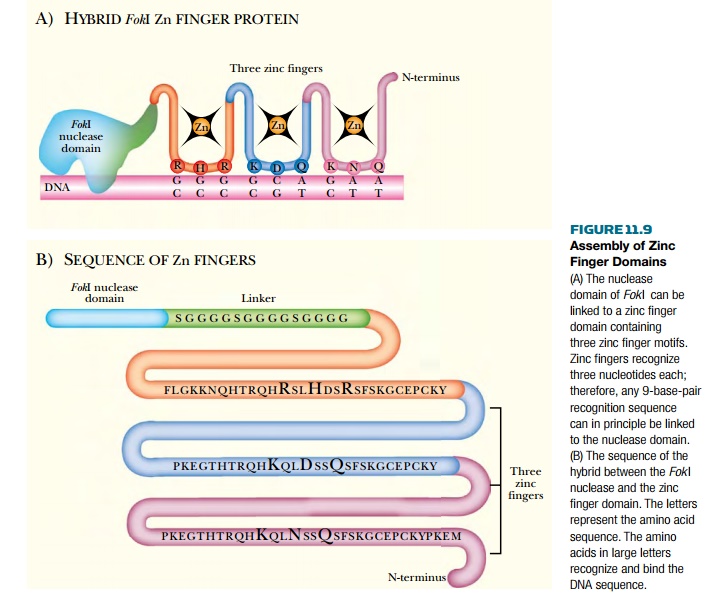
Zinc finger domains have also
been joined to the nuclease domain of FokI.
The zinc finger is a common DNA binding motif found in many regulatory
proteins. The zinc finger consists of 25 to 30 amino acids arranged around a Zn
ion, which is held in place by binding to conserved cysteines and histidines.
Each zinc finger motif binds three base pairs, and a zinc finger domain may
possess several motifs. In this example, domains approximately
90 amino acids long and
comprising three zinc finger motifs, and which therefore specifically recognize
nine base DNA sequences, were connected to the FokI nuclease domain (Fig. 11.9).
Both the amino acid sequence
and the corresponding DNA recognition specificity are now known for a wide
range of zinc fingers. This information allows zinc finger domains to be
designed to read any chosen DNA sequence. DNA segments that encode zinc finger
domains may be derived either from naturally occurring DNA binding proteins or
by artificial synthesis, because individual zinc finger motifs are about 25
amino acids (75 nucleotides) in length. In principle, engineered proteins can
now be provided with a zinc finger domain that will enable them to bind
specifically to any chosen DNA sequence.
Related Topics