Chapter: Biotechnology Applying the Genetic Revolution: Protein Engineering
Adding New Functional Groups Using Nonnatural Amino Acids
ADDING NEW FUNCTIONAL GROUPS USING NONNATURAL AMINO ACIDS
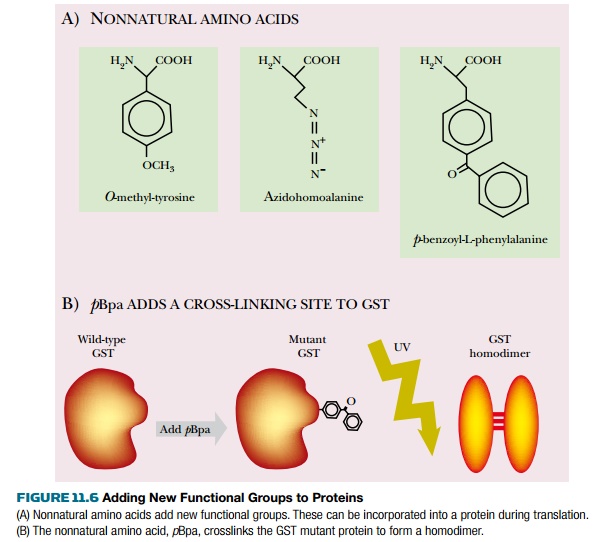
Many nonnatural amino acids
have different functional groups that are useful in protein engineering. For
example, adding p-benzoyl-L-phenylalanine (pBpa) into one position of glutathione-S-transferase (GST) adds a crosslinking
group that can be activated by UV irradiation. When GST is modified in this
way, UV irradiation creates a covalently linked homodimer (Fig. 11.6).
Incorporating a nonnatural amino acid into a protein can be done on a small scale by isolating the tRNA for a particular amino acid and attaching the nonnatural amino acid. The charged tRNA is then added to an in vitro protein translation system, which incorporates the nonnatural amino acid into the growing polypeptide chain. The chemical method is too costly and time consuming for large-scale use. For large-scale incorporation, E. coli may be modified to insert the nonnatural amino acid in vivo.
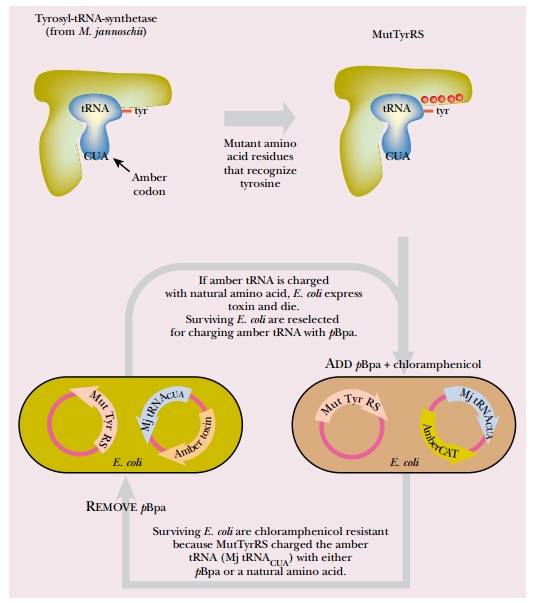
Inserting a nonnatural amino acid during in vivo protein synthesis requires a mutant aminoacyl-tRNA synthetase that charges a tRNA with the nonnatural amino acid. The laboratory of Peter G. Schultz, at the Scripps Research Institute, has developed an E. coli strain that incorporates pBpa at a specific amber codon. Directed evolution was used to mutate a tyrosyl-tRNA synthetase from M. jannaschii. The enzyme from M. jannaschii was used because it does not recognize any endogenous E. coli tRNA. Consequently, it needs the gene for its specific partner tRNA to be provided as well. In addition, the partner tRNA was altered so that it recognizes the amber stop codon instead of its original natural codon (i.e., it is an amber mutant tRNA). The result is that when the mutant tyrosyl-tRNA synthetase is expressed in E. coli, it inserts whichever amino acid it is charged with at amber stop codons.
To alter the tyrosyl-tRNA synthetase, a library of mutant enzymes was generated by random mutation of each amino acid residue involved in recognition of the amino acid substrate (originally tyrosine). The library of mutant tRNA synthetase genes was transformed into E. coli that possess a gene for the partner tRNA, and a gene for chloramphenicol resistance with an amber codon in the middle. The E. coli were grown in the presence of pBpa and chloramphenicol. If the mutant tRNA synthetase was able to insert an amino acid at the amber stop codon, then the chloramphenicol resistance gene (CAT) was expressed, and the cells lived. Otherwise, the cells died. This was the positive selection (Fig. 11.7).
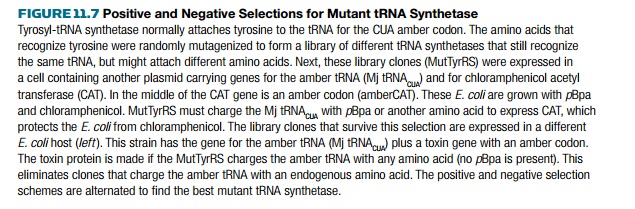
This positive selection does
not exclude mutant tRNA synthetases that charge the amber tRNA with a natural
amino acid, so a negative selection scheme was used next. The plasmids carrying
the mutant tRNA synthetases were isolated and transformed into a different E. coli strain. This E. coli had a toxin gene with an amber
suppressor mutation plus the amber tRNA. Here the mutants were grown without
any pBpa. If the mutant tRNA
synthetase could charge the amber tRNA with a natural amino acid, the toxin
would be made and the E. coli would
die. This eliminated mutant tRNA synthetases that used natural amino acids. The
selection scheme was repeated numerous times, and finally a specific mutant
tRNA synthetase was isolated (Fig. 11.7) that recognized the amber tRNA and
linked pBpa to it.
This altered tRNA synthetase
can be used to insert the nonnatural amino acid pBpa into other proteins, such as the GST mutant described earlier.
Thus it can be used to engineer crosslinking agents at any point in any target
protein. To achieve this, the researcher must insert an amber codon into the
gene that encodes the protein of interest and express the mutant gene in an E. coli host cell that expresses the
altered tRNA synthetase.
Related Topics