Chapter: Biotechnology Applying the Genetic Revolution: Protein Engineering
Changing Binding Site Specificity
CHANGING
BINDING SITE SPECIFICITY
In addition to altering the
overall stability of a protein, it is possible to deliberately change the
active site. The most straightforward alterations to make are those that change
the binding specificity for the substrate or a cofactor, but do not disrupt the
enzyme mechanism. Changing the specificity for a cofactor or substrate may be
useful, either to make the product of the enzyme reaction less costly or to
change it chemically.
This principle has been
demonstrated with several enzymes that use the cofactors NAD or NADP to carry
out dehydrogenation reactions. Both cofactors carry reducing equivalents and
both react by the same mechanism. Although a few enzymes can use either NAD or
NADP, most use one or the other. Generally, NAD is used by dehydrogenases in
degradative pathways, and the respiratory chain oxidizes the resulting NADH. In
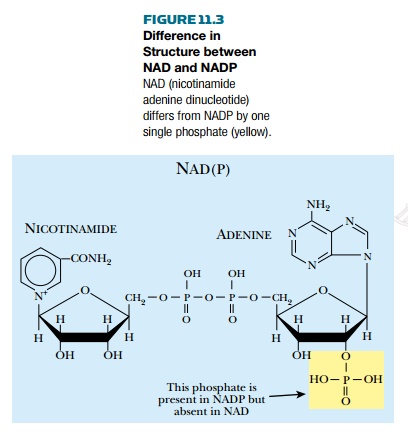
contrast, biosynthetic enzymes use NADP. Structurally they differ only in NADP
having an extra phosphate group attached to the ribose ring (Fig. 11.3). This
gives NADP an extra negative charge and, not surprisingly, enzymes that prefer
NADP have somewhat larger binding pockets with positively charged amino acid
residues at the bottom. Enzymes that favor NADH often have a negatively charged
amino acid residue in the corresponding position.
Several enzymes that use NAD
or NADP have been engineered to change their preference. For example, the
lactate dehydrogenase (LDH) of most bacteria uses reduced NAD, not NADP, to
convert pyruvate to lactate.
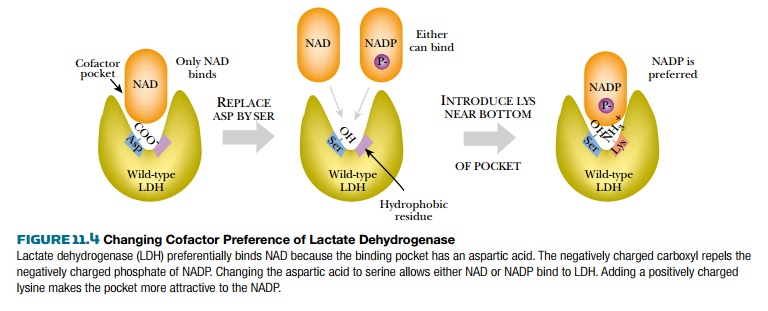
A conserved aspartate
provides the negative charge at the bottom of the cofactor binding pocket that
excludes NADP. If this is changed to a neutral residue, such as serine, the
enzyme becomes able to use both NADP and NAD. If, in addition, a nearby
hydrophobic residue in the cofactor pocket is replaced by a positively charged
amino acid (such as lysine or arginine), the enzyme now prefers NADP to NAD
(Fig. 11.4).
The specificity of LDH for its substrate can be altered in a similar way. The natural substrate lactate is a three-carbon hydroxyacid. It is possible to alter several residues surrounding the substrate binding site without impairing the enzyme reaction mechanism. By replacing a pair of alanines with glycines, the binding site can be made larger. By replacing hydrophilic residues (Lys, Gln) with hydrophobic ones (Val, Met), the site becomes more hydrophobic. Alteration of multiple residues gives an engineered LDH that accommodates five or six carbon analogs of lactate and uses them as substrates.
Related Topics