Chapter: Biotechnology Applying the Genetic Revolution: Protein Engineering
Engineering Disulfide Bonds
ENGINEERING
DISULFIDE BONDS
The formation of disulfide
bonds is a major factor in maintaining the 3D structure of many proteins.
Disulfide bonds are especially important for those proteins found outside the
cell in oxidizing environments. In practice, most enzymes used industrially
will be exposed to such oxidizing conditions, and therefore disulfide bonds are
particularly relevant.
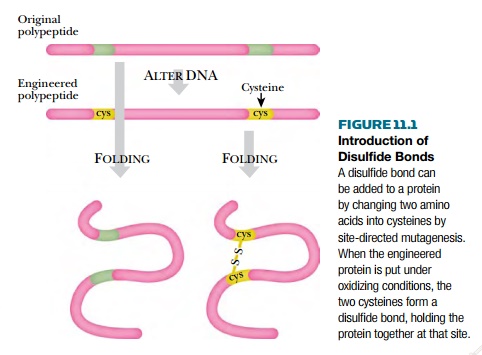
Introduction of extra
disulfide bonds is a relatively straightforward way to increase the stability
of proteins. The first step is to simply introduce two cysteine residues into
the polypeptide chain. Then, under oxidizing conditions, these will form a
disulfide bond provided that the polypeptide chain folds so as to bring the two
cysteines into close contact. Obviously, for this approach to work, the
tertiary structure of the protein must be known so that the cysteines can be
inserted in appropriate positions (Fig. 11.1). In general, the longer the loop
of amino acids between the two cysteines, the greater the increase in stability.
Note however, that formation of a disulfide linkage can create a strained
conformation if the two cysteines are not properly aligned. This may result in
a decrease in stability of the protein.
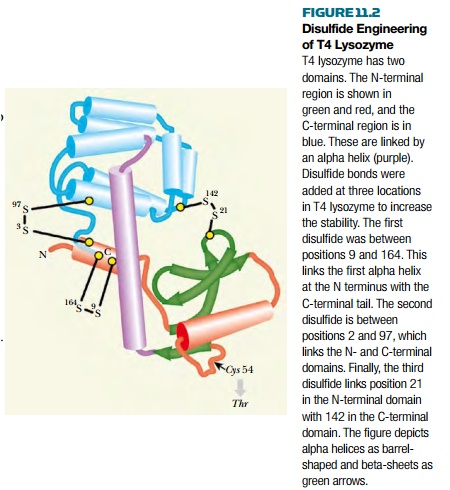
This approach has been
demonstrated using the enzyme lysozyme from bacterial virus T4. The structure
of this enzyme has been solved by x-ray crystallography. The polypeptide chain
of 164 amino acids folds into two domains and has two cysteines, neither
involved in disulfide bond formation in the wild-type protein. One of these,
Cys54, was first mutated to Thr to avoid formation of incorrect disulfides. The
other, Cys97, was retained for use in disulfide formation. Extensive analysis
of possible locations for disulfides was carried out. Those disulfides that
might impair other stabilizing interactions in the protein were eliminated.
This left three possible disulfide bonds that should theoretically promote
stability, located between positions 3 and 97, 9 and 164, and 21 and 142 (Fig.
11.2).
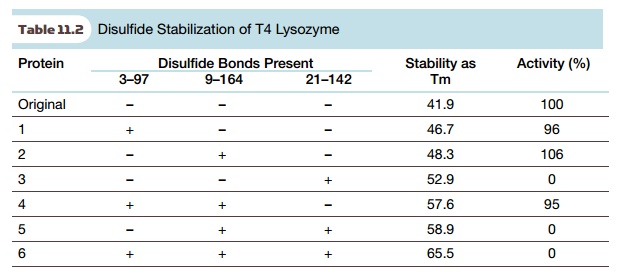
To test these experimentally,
five amino acids (Ile3, Ile9, Thr21, Thr142, and Leu164) were converted to Cys
in various combinations. Engineered proteins with each individual disulfide as
well as proteins with two or three disulfide linkages were tested for stability
and for enzyme activity (Table 11.2). Stability was measured by thermal
denaturation; the melting temperature,
Tm, is the temperature at which 50% of the protein is denatured. Although
all three disulfides increased stability of T4 lysozyme, those engineered
proteins with the 21–142 disulfide had lost their enzyme activity. The precise
reason is uncertain, but presumably some structural distortion has altered the
active site. Nonetheless, the engineered protein with both of the two other
disulfides showed a massive increase in stability and retained almost all of
its enzyme activity. Overall, this approach is highly effective.
Related Topics