Chapter: Biotechnology Applying the Genetic Revolution: Protein Engineering
Combinatorial Protein Libraries
COMBINATORIAL
PROTEIN LIBRARIES
So far we have discussed ways
to modify a useful protein that already exists. Another approach to protein
engineering is to generate large numbers of different protein sequences and
then screen them for some useful enzyme activity or other chemical property.
(Screening is often done by phage display or related techniques as described)
Rather than merely generating large numbers of random polypeptides, combinatorial screening usually uses
pre-made modules of some sort to create a random
shuffling library. For example, protein motifs known to provide a binding
site for metal ions or metabolites might be combined with segments known to
form structures such as an alpha helix.
In a common approach, DNA
modules of around 75 base pairs (i.e., 25 codons) are made by chemical DNA
synthesis. Several modules are then assembled to give a new artificial gene.
The modules are usually joined by PCR using overlapping primers Modules may be
joined in a chosen order or in a randomized manner. For proper expression of
the assembled sequence, the front and rear modules are normally specified to
provide suitable promoter and terminator sequences. The intervening modules may
then be randomly shuffled to generate more possible variation (Fig. 11.12).
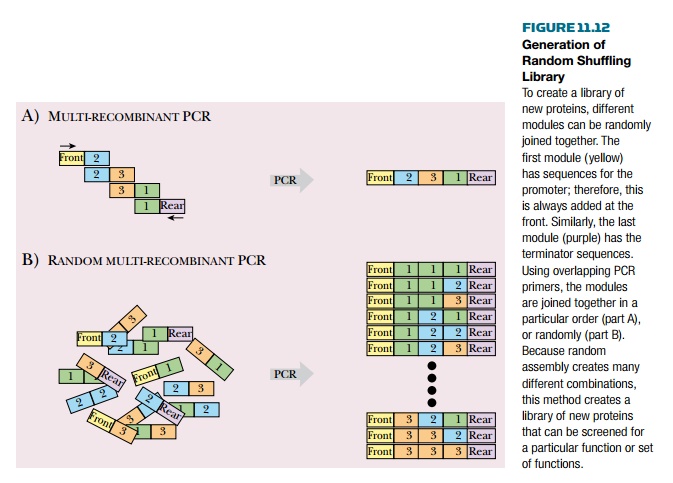
The resulting protein library
is then screened for some activity associated with the modules used. For
example, if modules for binding an organic metabolite and for binding Fe ions
were included, then the library of products might be screened for an enzymatic
activity that oxidizes the metabolite via iron-mediated catalysis.
A related approach is based
on the idea that the exons of eukaryotic genes encode modular segments of
proteins, such as binding sites and structural motifs. Although it is by no
means always true, in many cases exon boundaries do correspond to the ends of
structural domains within the encoded proteins. Consequently, it is believed
that at least some eukaryotic genes have evolved by the natural shuffling of
exons.
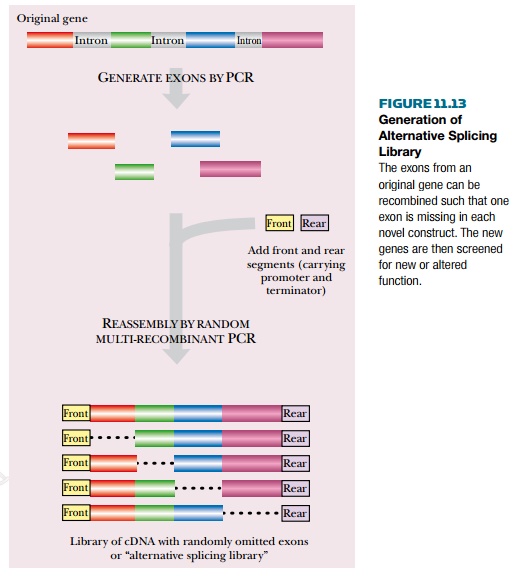
In the exon shuffling
approach a combinatorial library is
generated from an already existing eukaryotic gene. Each of the exons of the
eukaryotic gene is generated by a separate PCR reaction. The segments are then
mixed and reassembled by overlap PCR. Two variants exist, depending on the
design of the overlap primers for the PCR assembly. Reassembling the exons in
random order generates a random splicing library, much as described earlier.
Less radically, an alternative splicing
library retains the order of the exons but includes or excludes any
particular exon at random (Fig. 11.13). The final products are prescreened to
obtain sequences long enough to encode most of the original exons, as this
obviously greatly increases the chances of a functional protein.
Related Topics