Chapter: Biotechnology Applying the Genetic Revolution: Recombinant DNA Technology
Screening the Library of Genes by Hybridization
SCREENING
THE LIBRARY OF GENES BY HYBRIDIZATION
Once the library is
assembled, researchers often want to identify a particular gene or segment of
DNA within the library. Sometimes the gene of interest is similar to one from
another organism. Sometimes the gene of interest contains a particular
sequence. For example, many enzymes use ATP to provide energy. Enzymes that
bind ATP share
a common signature sequence
whether they come from bacteria or humans. This sequence can be used to find
other enzymes that bind ATP. Such common sequence motifs may also suggest that
a protein will bind various cofactors, other proteins, and DNA, to name a few
examples.
Screening DNA libraries by
hybridization requires preparing the library DNA and preparing the labeled
probe. A gene library is stored as a bacterial culture of E. coli cells, each having a plasmid with a different insert. The
culture is grown up, diluted, and plated onto many different agar plates so
that the colonies are spaced apart from one another. The colonies are
transferred to a nylon filter and the DNA from each colony is released from the
cells by lysing them with detergent. The cellular components are rinsed from
the filters. The DNA sticks to the nylon membrane and is then denatured to form
single strands (Fig. 3.20).
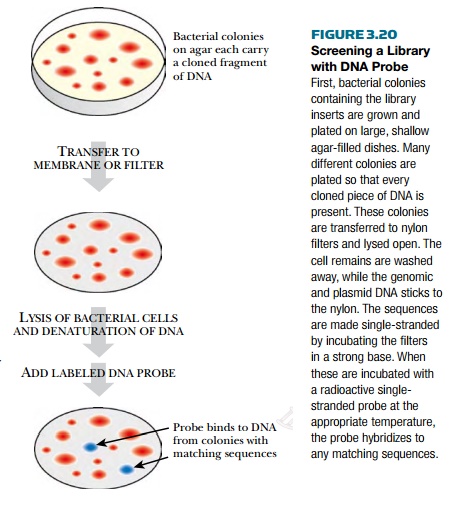
If a scientist is looking for
a particular gene in the target organism, the probe for the library may be the
corresponding gene from a related organism. The probe is usually just a segment
of the gene, because a smaller piece is easier to manipulate. The probe DNA can
be synthesized and labeled either with radioactivity or with chemiluminescence.
The probe is heated to make it single-stranded and mixed with the library DNA
on the nylon filters. The probe hybridizes with matching sequences in the
library. The level of match needed for binding can be adjusted by incubating at
various temperatures. The higher the temperature, the more stringent, that is,
the more closely matched the sequences must be. The lower the temperature, the
less stringent. If the probe is labeled with radioactivity, photographic film
will turn black where the probe and library DNA hybridized. The black spot is
aligned with the original bacterial colony. Usually the most likely colony plus
its neighbors are selected, grown, plated, and rescreened with the same probe
to ensure that a single transformant is isolated. Then the DNA from this
isolate can be analyzed by sequencing.
Related Topics