Chapter: Biotechnology Applying the Genetic Revolution: Recombinant DNA Technology
Hybridization of DNA or RNA in Southern and Northern Blots
HYBRIDIZATION
OF DNA OR RNA IN SOUTHERN AND NORTHERN BLOTS
If two different double
helixes of DNA are melted, the single strands can be mixed together before
cooling and reannealing. If the two original DNA molecules have similar
sequences, a single strand from one may pair with the opposite strand from the
other DNA molecule.
This is known as hybridization and can be used to
determine whether sequences in two separate samples of DNA or RNA are related.
In hybridization experiments, the term probe
molecule refers to a known DNA
sequence or gene that is used to screen the experimental sample or target DNA
for similar sequences.
Southern blots are used to determine how closely DNA from one source is related to
a DNA sequence from another source.
The technique involves forming hybrid DNA molecules by mixing DNA from the two
sources. A Southern blot has two components, the probe sequence (e.g., a known
gene of interest from one organism) and the target DNA (often from a different
organism). A typical Southern blot begins by isolating the target DNA from one
organism, digesting it with a restriction enzyme that gives fragments from
about 500 to 10,000 base pairs in length, and separating these fragments by
electrophoresis. The separated fragments will be double-stranded, but if the
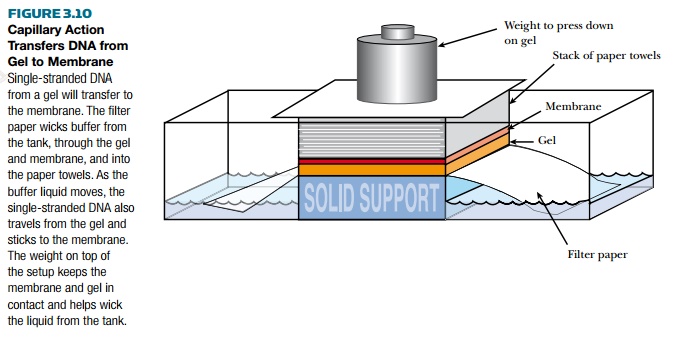
gel is incubated in a strong acid, the DNA separates into single strands. Using
capillary action, the single strands can be transferred to a membrane as shown
in Fig. 3.10. The DNA remains single-stranded once attached to the membrane.
Next, the probe is prepared.
First, the known sequence or gene must be isolated and labeled in some way (see
earlier discussion). Identifying genes has become easier now that many genomes
have been entirely sequenced. For example, a scientist can easily obtain a copy
of a human gene for use as a probe to find similar genes in other organisms.
Alternatively, using sequence data, a unique oligonucleotide probe can be designed
that only recognizes the gene of interest. If an oligonucleotide has a common
sequence, it will bind to many other sequences. Therefore, oligonucleotide
probes must be long enough to have sequences that bind to only one (or very
few) specific site(s) in the target genome. To prepare DNA probes for a
Southern blot, they are labeled using radioactivity,
biotin, or digoxigenin (see
earlier discussion). Finally, the labeled DNA is denatured at high temperature
to make it single-stranded. (Synthetic oligonucleotides do not require
treatment, as they are already single-stranded.)
To perform the Southern blot,
the single-stranded probe is incubated with the membrane carrying the
single-stranded target DNA (Fig. 3.11). These are incubated at a temperature
that allows
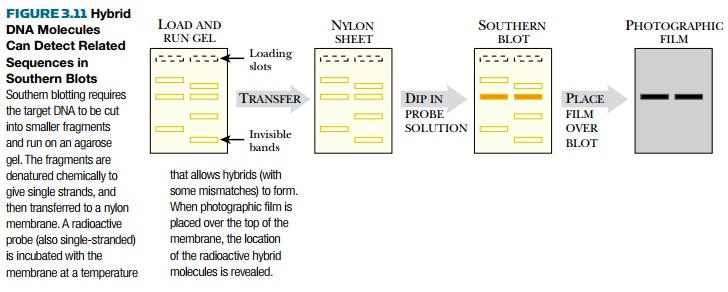
hybrid DNA strands to form
but allows a low amount of mismatching. The temperature, and hence the level of
mismatching tolerated, can be varied depending on how specific a search is
being run. If the probe is radioactive, then the membrane is exposed to
photographic film. If the probe is labeled with biotin or digoxigenin, the
membrane may be treated with chemiluminescent substrate to detect the labeled
probe, and then exposed to photographic film. Dark bands on the film reveal the
positions of fragments with similar sequence to the probe. Alternatively,
biotin or digoxigenin labels may be visualized by treatment with a chromogenic
substrate. In this case blue bands will form directly on the membrane at the position
of the related sequences.
Northern blots are also based on nucleic acid hybridization. The difference is
that RNA is the target in a Northern
blot. The probe for a Northern blot is either a fragment of a gene or a unique
oligonucleotide just as in a Southern blot. The target RNA is usually messenger
RNA. In eukaryotes, screening mRNA is more efficient because genomic DNA has a
lot of introns, which may interfere with probes binding to the correct
sequence. Besides, mRNA is already single-stranded, so the agarose gel does not
have to be treated with strong acid. Much like a Southern blot, Northern blots
begin by separating mRNA by size using electrophoresis. The mRNA is transferred
to a nylon membrane and incubated with a single-stranded labeled probe. As
before, the probe can be labeled with biotin, digoxigenin, or radioactivity.
The membrane is processed and
exposed to film or chromogenic substrate.
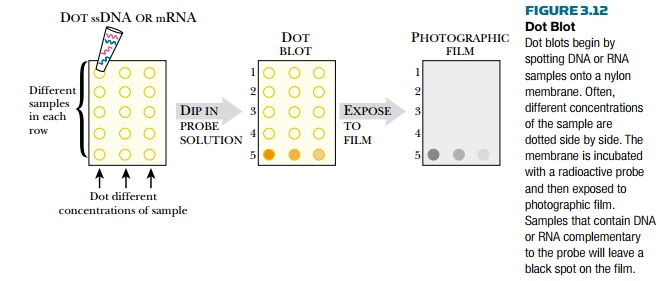
A variation of these
hybridization techniques is the dot blot (Fig. 3.12). Here the target sample is
not separated by size. The DNA or mRNA target is simply attached to the nylon
membrane as a small dot. As in Southern blots, the DNA sample must be made
single-stranded before it is attached to the membrane. As before, the dot-blot
membrane is allowed to hybridize with a labeled probe. The membranes are
processed and exposed to film. If the dot of DNA or mRNA contains a sequence
similar to the probe, the film will turn black in that area. Dot blots are a
quick and easy way to determine if the target sample has a related sequence,
before more detailed analysis by Southern or Northern blotting. Another
advantage of dot blots is that multiple samples can be processed in a smaller
amount of space.
Related Topics