Chapter: Biotechnology Applying the Genetic Revolution: Recombinant DNA Technology
Methods of Detection for Nucleic Acids
METHODS OF DETECTION FOR NUCLEIC ACIDS
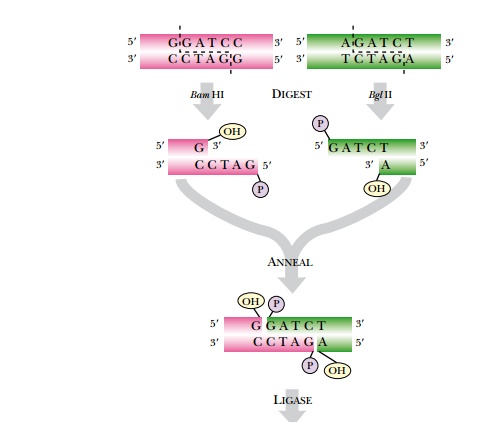
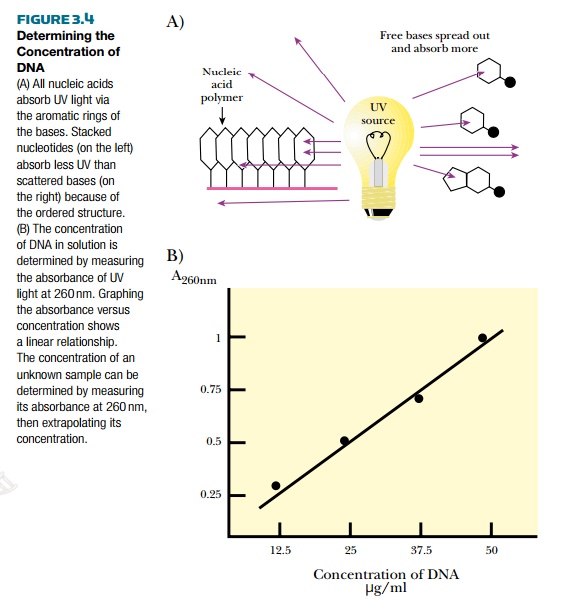
Recombinant DNA methodologies require the ability to detect DNA. One of the easiest ways to detect the amount of DNA or RNA in solution is to measure the absorbance of ultraviolet light (Fig. 3.4). DNA absorbs ultraviolet light because of the ring structures in the bases.
Single-stranded RNA and free nucleotides also absorb ultraviolet light. In fact, they absorb more light because their structures are looser. The amount of absorption is compared with a known set of standards and the concentration of DNA can be determined.
Radioactive Labeling of Nucleic Acids and Autoradiography
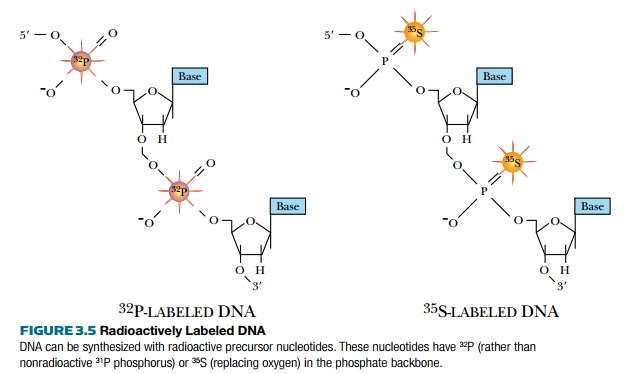
Ultraviolet light absorption is a general method for detecting DNA, but does not distinguish between different DNA molecules. DNA can also be detected with radioactive isotopes
(Fig. 3.5). During replication, radioactive precursors such as 32P in the form of a phosphate group and 35S in the form of phosphorothioate can be incorporated. Because native DNA does not contain sulfur atoms, one of the oxygen atoms of a phosphate group is replaced with sulfur to make phosphorothioate. Most radioactive molecules used in laboratories are short lived. 32P has a half-life of 14 days and 35S has a half-life of 68 days, so the isotopes degrade fairly fast. Although radioactive DNA is invisible, photographic film will turn black when exposed to the radioactive DNA. Radioactively labeled DNA is considered “hot,” whereas unlabeled DNA is considered “cold.”
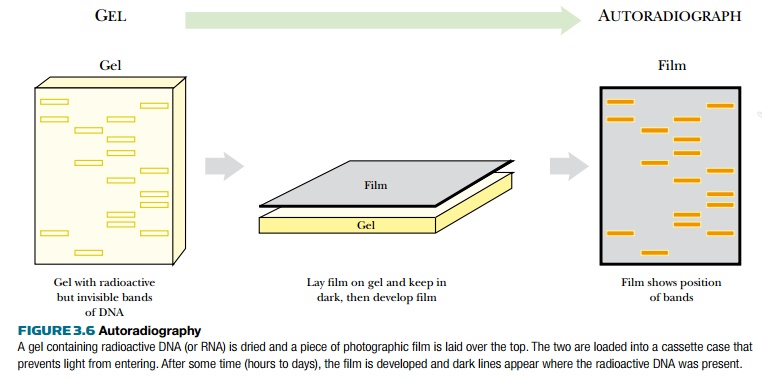
The radioactive nucleotide precursors can be supplied to rapidly growing bacterial cultures. During replication, the radioactive precursor is incorporated into new DNA. The DNA is isolated from the bacteria and run on a gel. Autoradiography identifies the location of radioactively labeled DNA in the gel (Fig. 3.6). If the gel is thin, like most polyacrylamide gels, it is dried with heat and vacuum. If the gel is thick, like agarose gels, the DNA is transferred to a nylon membrane using capillary action (see Fig. 3.9, later). The dried gel or nylon membrane is placed next to photographic film. As the radioactive phosphate decays, the radiation turns the photographic film black. Only the areas next to radioactive DNA will have black spots or bands. The use of film detects where the hot DNA is on a gel, and the use of ethidium bromide shows where all of the DNA, hot or cold, is. These two methods allow distinguishing one DNA fragment from another.
Radioactive DNA can also be detected using scintillation counting. Here a small sample of the radioactive DNA is mixed with scintillation fluid. When the radioactive isotope decays, it emits a beta particle. Scintillation fluid emits a flash of light when excited by the beta particle. The scintillation counter detects light flashes with a photocell, counting them over a specified amount of time. Radioactive DNA concentrations can be determined by comparing to a set of known standards. Scintillation counting cannot detect the unlabeled or cold DNA, nor can it distinguish between multiple fragments of hot DNA, because it merely measures the total radioactivity in the sample.
Fluorescence Detection of Nucleic Acids
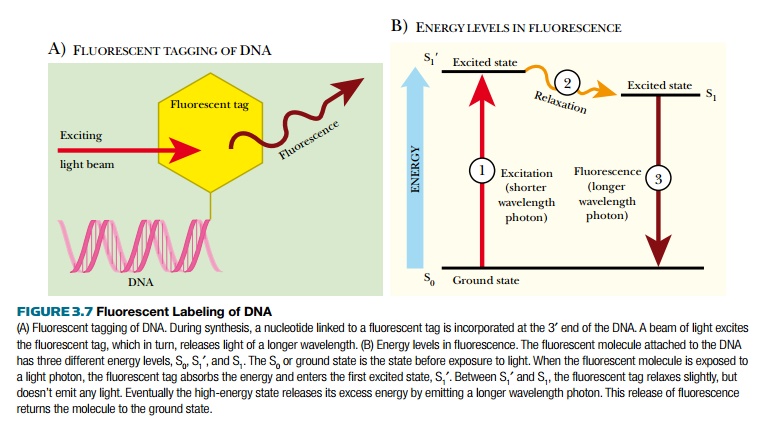
Autoradiography has its merits, but working with and disposing of radioactive waste is costly, both monetarily and environmentally. Using fluorescently tagged nucleotides was developed as a better method of DNA detection (Fig. 3.7). Fluorescent tags absorb light of one wavelength, which excites the atoms, increasing the energy state of the tag. This excited state releases a photon of light at a different (longer) wavelength and returns to the ground state. The emitted photon is detected with a photodetector. There are many different fluorescent tags, and each emits a different wavelength of light. Some photodetector systems are sensitive enough to distinguish between these different tags; therefore, if different bases have different fluorescent labels, the photodetector can determine which base is present. This is the basis for most modern DNA sequencing machines.
Chemical Tagging with Biotin or Digoxigenin
Biotin is a vitamin and digoxigenin is a steroid from the foxglove plant. Using these two chemicals allows scientists to label DNA without radioactivity or costly photodetectors. Biotin or digoxigenin are chemically linked to uracil; therefore, DNA must be synthesized with the labeled uracil replacing thymine. The DNA is synthesized in vitro as described. A single-stranded DNA template, DNA polymerase, a short DNA primer, and a mixture of dATP, dGTP, dCTP, plus dUTP linked to either biotin or digoxigenin are combined. DNA polymerase synthesizes the complementary strand to the template, incorporating biotin- or digoxigenin-linked uracil opposite all the adenines.
The labeled DNA is visualized in a two-step process (Fig. 3.8). First, for biotin, a molecule of avidin binds to the tag. For digoxigenin, a specific antibody binds to the tag. Both avidin and the digoxigenin antibody are conjugated to alkaline phosphatase, an enzyme that
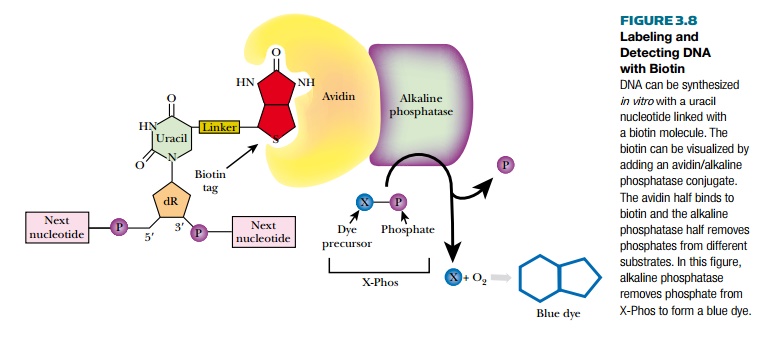
removes phosphates from a variety of substrates. Several different chromogenic molecules act as substrates for alkaline phosphatase, but the most widely used one is X-Phos. Once alkaline phosphatase removes the phosphate group from X-Phos, the intermediate molecule reacts with oxygen and forms a blue precipitate. This blue color reveals the location of the labeled DNA. Another substrate of alkaline phosphatase is Lumi-Phos, which is chemiluminescent and emits visible light when the phosphate is removed. Much like autoradiography, when photographic film is placed over labeled DNA treated with Lumi-Phos, dark bands form wherever the Lumi-Phos glows.
Related Topics