Chapter: Biotechnology Applying the Genetic Revolution: Recombinant DNA Technology
Eukaryotic Expression Libraries
EUKARYOTIC
EXPRESSION LIBRARIES
In expression libraries, the vector has sequences required for
transcription and translation of the insert. This means that the insert DNA is
expressed as RNA, and this may be translated into a protein. An expression
library, in essence, generates a protein from every cloned insert, whether it
is a real gene or not. When studying eukaryotic DNA, expression libraries are
constructed using complementary DNA
(cDNA) to help ensure the insert is truly a gene. Eukaryotic DNA,
especially in higher plants and animals, is largely noncoding, with coding
regions spaced far apart. Even genes are interrupted with noncoding introns.
cDNA is a double-stranded DNA copy of mRNA made by using reverse transcriptase.
Reverse transcriptase was first identified in retroviruses. It is used in
eukaryotic research to eliminate the introns and generate a version of a gene
consisting solely of an uninterrupted coding sequence.
In contrast, bacteria have
very little noncoding DNA and their genes are not interrupted by introns;
therefore, genomic DNA can be used directly in expression libraries.
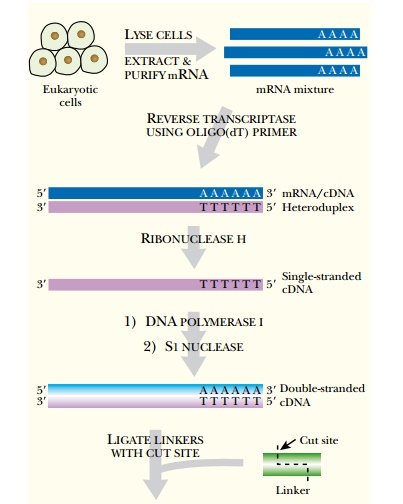
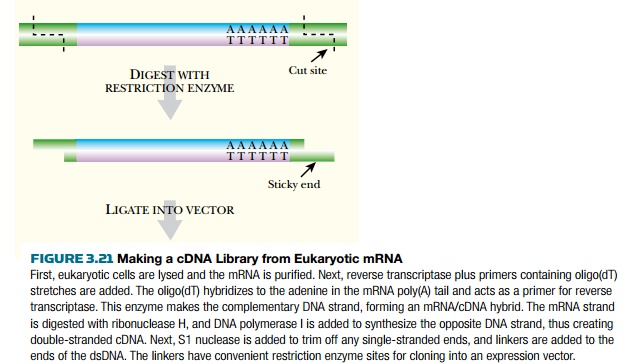
Eukaryotic DNA is first made into cDNA in order to construct an expression library (Fig. 3.21). To make cDNA, the messenger RNA is isolated from the organism of interest by binding to a column containing poly(T) (i.e., a DNA strand consisting of repeated thymines). This isolates only mRNA because poly(T) anneals to the poly(A) tail of eukaryotic mRNA. The mRNA is made double-stranded using reverse transcriptase, which synthesizes a DNA complement to mRNA. The hybrid mRNA:DNA molecule is converted to double-stranded DNA using RNase H and DNA polymerase I. RNase H cuts nicks into the mRNA backbone, and DNA polymerase displaces the mRNA strand and synthesizes DNA (see Fig. 3.21).
The cDNA is then ligated into
an expression vector with sequences
that initiate transcription and translation of the insert. In some cases, the
insert will have its own translation start site (e.g., a full-length cDNA). If,
as often occurs, the insert does not contain a translation start, then the
reading frame becomes an issue. Because the genetic code is triplet, each
insert can be translated in three different reading frames. A protein may be
produced for all three reading frames, but only one frame will actually produce
the correct protein. To ensure obtaining inserts with the correct reading
frame, each cDNA is cloned in all three reading frames by using linkers with
several different restriction sites. The number of transformants to screen is
therefore increased greatly.
The cloned genes are transformed
into bacteria, which express the foreign DNA. The bacteria are grown on agar
and the colonies are then transferred to a nylon membrane and lysed. The
proteins released are attached to the nylon membranes and are screened in
various ways.
Most often, an antibody to
the protein of interest is used. This recognizes the protein and can be
identified using a secondary antibody that is conjugated to a detection system.
Usually alkaline phosphatase is conjugated to the secondary antibody. The whole
complex can be identified because alkaline phosphatase cleaves X-Phos, leaving
a blue color where the bacterial colony expressed the right protein (Fig.
3.22). E. coli cannot perform most of
the posttranslational modifications that eukaryotic proteins often undergo.
Therefore, the proteins are not always in their native form. Nonetheless,
appropriate antibodies can detect most proteins of interest.
Related Topics