Botany : Chromosomal Basis of Inheritance - Protein synthesis in plants | 12th Botany : Chapter 3 : Chromosomal Basis of Inheritance
Chapter: 12th Botany : Chapter 3 : Chromosomal Basis of Inheritance
Protein synthesis in plants
Protein
synthesis in plants
The process of protein
synthesis consists of two major steps, they are Transcription and Translation.
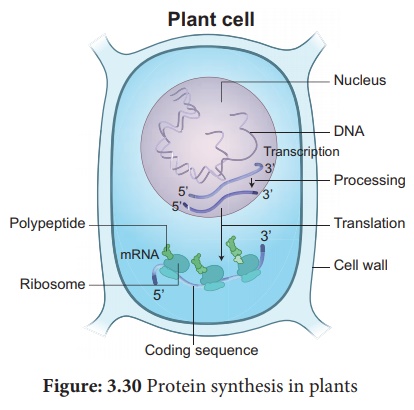
1. Transcription
Transcription is the
process in which one strand of DNA acts as a template to generate mRNA with the
bases complementary to the template strand. It is catalyzed by the enzymes
called RNA polymerases.
Transcription and processing of RNA
takes place in the nucleus, whereas the translation occurs in
the ribosomes found in cytoplasm. In Eukaryotes, mRNA molecules are
monocistronic with only one protein being derived from each mRNA.
The transcription begins
with unwinding of DNA double helix and the hydrogen bonds are broken at the
site of the gene being transcribed.
Template Strand /
Non-Coding Strand / Antisense Strand
The strand of DNA which
is oriented in 3’ → 5’ direction that serves as a template for the synthesis of mRNA
is called template strand.
Coding strand /
Non-template strand / Sense Strand
The other strand of DNA
which is not transcribed is called the Coding Strand.
A specific sequence of
DNA nucleotides called the Promoter is necessary for transcription to
takes place. It consists of TATA box and transcription start site where
transcription begins.
Termination sequences are the DNA sequences
which tells when the RNA polymerase should stop producing RNA molecule.
Eukaryotic structural
gene has 3 features in promoter
1. Regulatory elements
2. TATA box
3. A transcriptional start
site
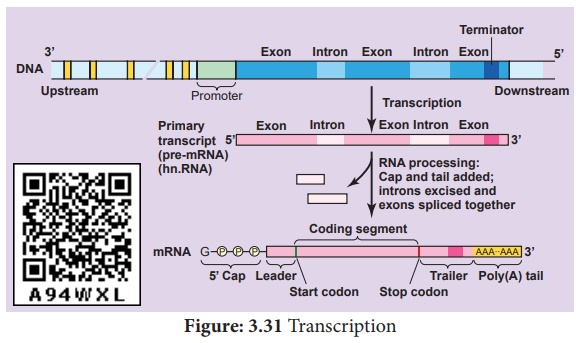
The transcription start site contains about 25 bp (basepairs) upstream, the sequence is TATAAT known as TATA or Hogness box which is present in core promoter. General transcriptional factors are the proteins which recognise base sequences of DNA and controls transcription. Some transcription factors bind directly to the promoter.
Some transcription
factors recognize the regulatory elements and bind to them to increase the rate
of transcription, others inhibits transcription.
To start the process of
transcription the Regulatory elements help the RNA polymerase to recognize core
promoter. The two categories of regulatory elements are
1. Enhancer sequences – they are DNA sequences
(activating sequences) which help to influence transcription.
2. Silencer sequence – DNA sequences that
inhibit transcription or decrease transcription.
Consensus sequence – An ideal sequence
in which each position represents the base which is found most often.
In addition to General
transcription factors (GTF) and RNA Pol II, a mediator is required for
transcription. The interactions between RNA polymerase II and regulatory TF
that bind to enhancers or silencers are mediated by a mediator.
RNA Polymerases cannot bind directly to the DNA, first it binds to the transcription factor which recognizes the promoter sequences which helps to find the protein coding regions of DNA.
RNA Polymerase with the
promoter sequence will transcribe the gene. Transcription factor plays an
important role in guiding RNA Polymerases to the promoter sequence. RNA
Polymerases bind RNA nucleotides together forming a growing strand in the 5’ → 3’ direction.
Transcription occurs in 5’ → 3’ direction, RNA Polymerase catalyses the addition of
nucleotides at the 3’ end of the growing chain of RNA.
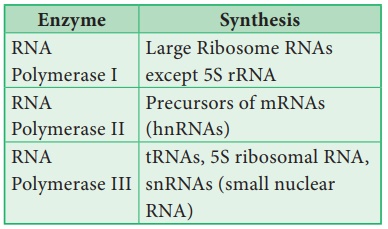
In Eukaryotes, 3
different RNA Polymerases called RNA Polymerases I, II and III are found.
The processing of pre-mRNA to mature mRNA / Molecular mechanism of RNA modification
In eukaryotes three
major types of RNA, mRNA, tRNA and rRNA are produced from a
precursorRNAmoleculetermedastheprimary transcript or preRNA. The RNA polymerase
II transcribes the precursor of mRNA, which are also called the heterogenous
nuclear RNA or hnRNA which are processed in the nucleus
before they are transported into the cytoplasm.
Capping
Modification at the 5’ end of the primary RNA transcript (hn RNA) with methylguanosine triphosphate is called capping.
Purpose of Capping
1. Protects RNA from
degradation.
2. Capping plays an
important role in removal of first intron in pre mRNA.
3. It regulates the mRNA
export from the nucleus into the cytoplasm.
4. It helps in binding of
mRNA to the ribosome.
Tailing / Polyadenylation
The 3’ end of hnRNA is
cleaved by an endonuclease and a string of adenine nucleotides is added to the
3’ end of hnRNA (pre mRNA) is known as Poly (A) tail - Polyadenylation. This
process is called tailing or polyadenylation.
Purpose of Tailing
1. Translation of RNA
transcript is facilitated.
2. Helps in the
synthesis of Polypeptides.
3. It enhances the mRNA
stability in the cytoplasm.
The protein coding
regions are not continuous in eukaryotes. Split genes were independently
discovered by Richard J Roberts and Phillip A. Sharp in 1977 and was awarded
Nobel Prize in 1993. Exons are the coding sequences or expressed
sequences contain biological informations in the matured processed mRNA. Introns
are intervening sequences, which are non-coding sequences (non
-amino acid-coding sequences) that should be removed from a gene before the
mRNA product is made. Introns do not code for any enzyme or structural protein
or polypeptides. These exons and introns are known as Split Genes.
2. RNA Splicing in plants
RNA Splicing is a
process which involves the cutting or removing out of introns and knitting of
exons. This process takes place in spherical particles which is a multiprotein
complex called SPLICISOMES. It is approximately 40 – 60 nm in diameter.
The spliceosomes have many small nuclear ribonucleic acids (snRNAs) and small
nuclear ribonuclear protein particles (snRNPs) which identify and helps in the
removal of introns.
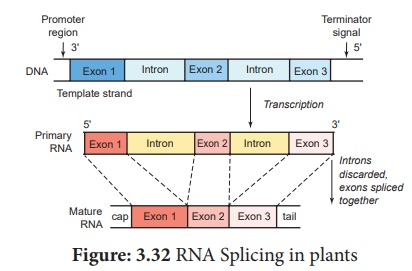
A spliceosome removes
the introns with an enzyme ribozyme. Now the mature mRNA comes away from the
spliceosomes through the nuclear pore and is transported out from the nucleus
into cytoplasm, and gets attached to ribosome to carry out translation. The RNA
and many proteins are transported through a nuclear pore complex by an energy
dependent process.
3. Translation
The genetic information
in the DNA code is copied onto mRNA bound in ribosomes for making polypeptides.
The mRNA nucleotide sequence is decoded into amino acid sequence of the protein
which is catalyzed by the ribosome. This process is called translation.
Terminology in Protein
synthesis
Codon – DNA codes are referred
to as triplet codes and those in mRNA is called as Codons. Each triplet
specifies a particular amino acid. Codons present in mRNA are read in 5’ → 3’ direction. There are
64 codons of which 61 codons codes for amino acids.
Start codon – AUG specified
methionine
Stop or Termination
codon –
UAA – Ochre UAG – amber and UGA – Opal.
Anticodons – The triplet of bases
in a tRNA molecule is known as anticodon. In tRNA sequence of three
bases which is complementary to codons of mRNA are called anticodon. The codons
of mRNA are recognized by the anticodons of tRNA which are oriented in 3’ → 5’ direction.
Process of translation
The following are major
steps in translation process
1. Initiation
The translation begins
with the AUG codon (start codon) of mRNA. Translation occurs on the surface of
the macromolecular arena called the ribosome. It is a nonmembranous organelle.
During the process of translation the two subunits of ribosomes unite (combine)
together and hold mRNA between them. The protein synthesis begins with the
reading of codons of mRNA. The tRNA brings amino acid to the ribosome, a
molecular machine which unites amino acids into a chain according to the
information given by mRNA. rRNA plays the structural and catalytic role during
translation.
A ribosome has one
binding site for mRNA and two for tRNA. The two binding sites of tRNA are
i.
P-Site – The peptidyl – tRNA binding site is one of the tRNA binding
site. At this site tRNA is held and linked to the growing end of the
polypeptide chain.
ii. A-Site – The
Aminoacyl – tRNA binding site. This is another tRNA binding site which
holds the incoming amino acids called aminoacyl tRNA. The anticodons of tRNA
pair with the codons of the mRNA in these sites.
2. Elongation of polypeptide chain
The P and A sites are
nearby, so that two tRNA form base pairs with adjacent codon. The polypeptide
chain is formed by the pairing of codons and anticodons according to the
nucleotide sequence of the mRNA.
Translators of the genetic code – tRNA
The tRNA translates the
genetic code from the nucleic acid sequence to the amino acid sequence i.e from
gene – Polypeptide. When an amino acid is attached to tRNA it is called aminoacylated
or charged. This is an energy requiring process which
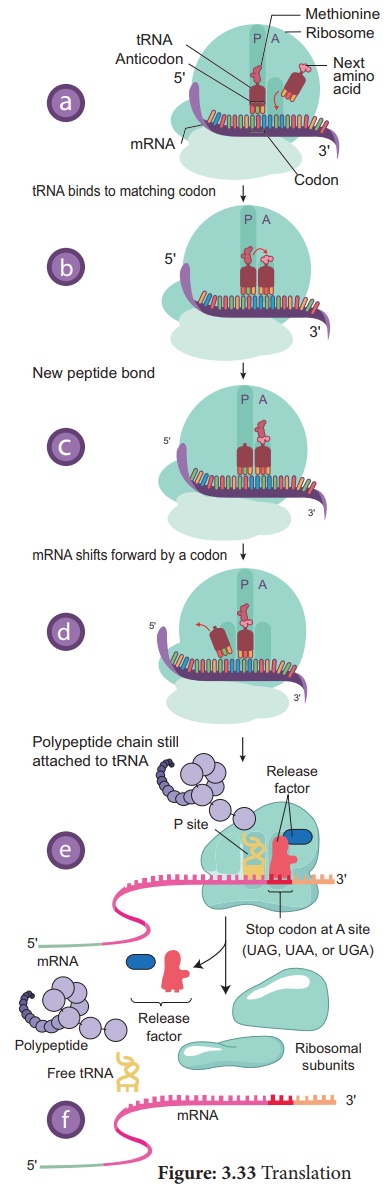
The translation begins
with the AUG codon (start codon) of mRNA. The tRNA which carries first amino
acid methionine attach itself to P-site of ribosome. The ribosome adds
new amino acids to the growing polypeptides. The second tRNA molecules has
anticodons which carries amino acid alanine pairs with the mRNA codon in
the A-site of the ribosome. The aminoacids methionine and alanine are
close enough so that a peptide bond is formed between them.
The bond between the
first tRNA and methionine now breaks. The first tRNA leaves the
ribosome and the P-site is vacant. The ribosome now moves one codon along the
mRNA strand. The second t-RNA molecule now occupies the P-site. The third t-RNA
comes and fills the A site (serine). Now a peptide bond is formed between alanine
and serine. The mRNA then moves through the ribosome by three
bases. This expels deacylated / uncharged tRNA from P-site and moves peptidyl
tRNA into the P- site and empties the A-site.This movement of tRNA from A-site
to P-site is said to be translocation. The translocation requires the hydrolysis
of GTP.
The ribosome (ribozyme -
peptidyl transferase) catalyses the formation of peptide bond by adding amino
acid to the growing polypeptide chain.
The ribosome moves from
codon to codon along the mRNA in the 5’ to 3’ direction. Amino acids are added one
by one translated into polypeptide as dictated by the mRNA. Translation is an
energy intensive process. A cluster of ribosomes are linked together by a
molecule of mRNA and forming the site of protein synthesis is called as polysomes
or polyribosomes.
3. Termination of polypeptide synthesis
Eukaryotes have
cytosolic proteins called release factors which recognize the termination
codon, UAA, UAG, or UGA when it is in the A site. When the ribosome reaches a
stop codon the protein synthesis comes to an end. So ribosomes are the protein
making factories of a cell. When the polypeptide is completed the ribosome
releases the polypeptide and detaches from the mRNA molecule. Now the ribosome
splits into small and large subunits after the release of mRNA.
4. Alternative Splicing in plants
It is very useful in
regulating gene expression to overcome the environmental stress in plants.
Alternative splicing is
an important mechanism / process by which multiple mRNA’s and multiple proteins
products can be generated from a single gene. The different proteins generated
are called isoforms. There are various modes of alternative splicing.
When multiple introns
are present in a gene, they are removed separately or as a unit. In certain
cases one or more exons which is present between the introns are also removed.
Significance of alternative splicing
1. The proteins transcribed from alternatively spliced mRNA containing different amino acid sequence lead to the generation of protein diversity and biological functions.
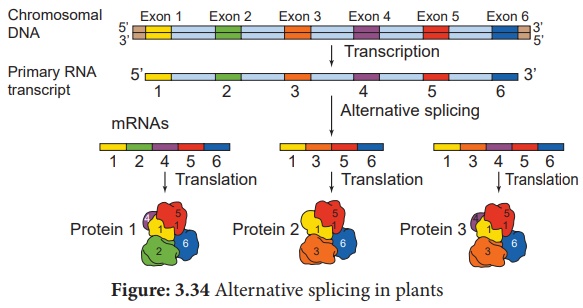
2. Multiple protein
isoforms are formed.
3. It creates multiple
mRNA transcripts from a single gene. A process of producing related proteins
from a single gene thereby the number of gene products are increased.
4. It plays an important
role in plant functions such as stress response and trait selection.
The plant adapts or
regulates itself to the changing environment.
5. RNA Editing – Post Transcriptional RNA Processing in plants
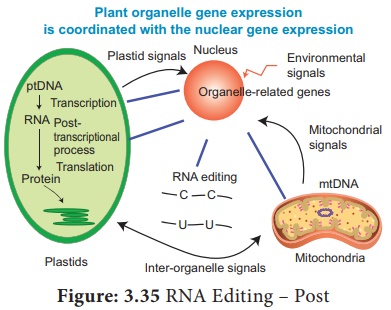
Figure: 3.35 RNA Editing
– Post
Transcriptional RNA
Processing in plants Chemical modification such as base modification,
nucleotide insertion or deletions and nucleotide replacements of mRNA results
in the alteration of amino acid sequence of protein that is specified is called
RNA editing.
This results in the
change in the protein coding sequence of RNA following transcription.
The coding properties of the RNA transcript is changed. The genetic information encoded in the chloroplast genome is altered by post transcriptional phenomenon which is site – specific (C → U) in chloroplast of higher plants – RNA editing occurs in plant mitochondria and chloroplast.
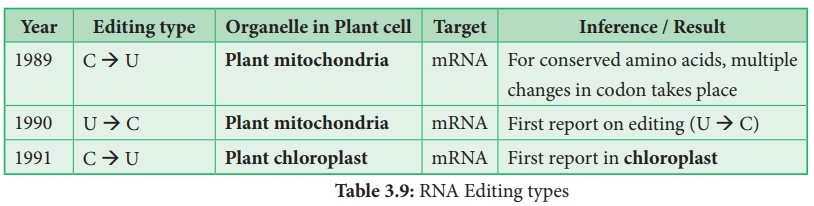
In plant cells RNA
editing by pyrimidine transitions occurs in mitochondria and plastids
(chloroplast) . There are two main types of RNA editing. (1) Substitution
editing – Alteration of individual nucleotide bases. Mitochondria and
chloroplast RNA in plants.
(2) Insertion / Deletion editing – Nucleotides are
added or deleted from the total number of bases.
Significance of RNA
Editing
1. In higher plant
chloroplast, it helps to restore the codons for conserved amino acids which
include initiation and termination codon.
2. It regulates Organellar
gene expression in plants.
3. RNA editing results in
the restoration of codons for phylogenetically conserved amino acid residues.
6. Jumping Genes

Have you heard of Jumping Genes or Hopping Genes?
This is the nick name of
transposable genetic elements. Transposons are the DNA sequences which can move
from one position to another position in a genome. This was first reported in
1948 by American Geneticist Barbara McClintock as “mobile controlling element”
in Maize. One of the most significant scientists of 20th century was Barbara McClintock
because she gave a shift in gene organization. McClintock was awarded Nobel
prize in 1983 for her work on transposons. Barbara McClintock when studying
aleurone of single maize kernels, noted the unstable inheritance of the mosaic
pattern of blue, brown and red spots due to the differential production of
vacuolar anthocyanins.
In maize plant genome
has AC / Ds transposon (AC = Activator, Ds = Dissociation). The activity of AC
element is very distinct in maize plant. The transposition in somatic cells
results in the changes in gene expression such as variegated pigmentation in
maize kernels. Maize genome has transposable elements which regulated the
different colour pattern of kernels.
McClintock’s findings
concluded that Ds and AC genes were mobile controlling elements. We now call it
as transposable elements, a term coined by maize geneticist, Alexander Brink.
McClintock gave the first direct experimental evidence that genomes are not
static but are highly plastic entities.
Significance of transposons
1. They contribute to many
visible mutations and mutation rate in an Organism.
2. In evolution, they
contribute to genetic diversity.
3. In genetic research
transposons are valuable tools which are used as mutagens, as cloning tags,
vehicles for inserting foreign DNA into model organism.
Plant genome – The word genome is
defined as the full complement of DNA (including all the genes and the
intergenic regions) present in an organism It specifies the entire biological
information of an organism. There are three distinct genomes in eukaryotic
cells and they are (1) The nuclear genome (2) The mitochondrial genome and (3)
The chloroplast genome present only in plants.
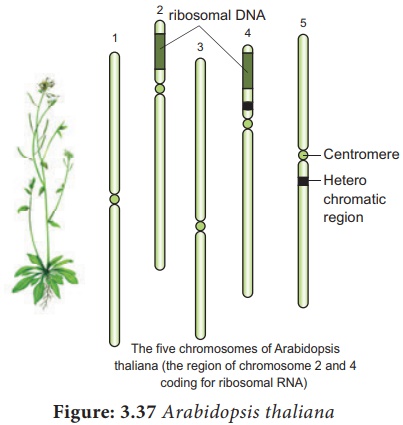
Arabidopsis thaliana – Thale cress, Mouse-ear
cress
1. It is a model plant for
the study of genetic and molecular aspects of plant development.
2. It belongs to mustard
family and it is the first flowering plant, where its entire genome is
sequenced.
3. The two regions of the
nucleolar organiser ribosomal DNA which codes for the ribosomal RNA are present
at the extremity of chromosomes 2 and 4
4. It is Diploid plant
having small genome with 2n = 10 chromosomes. Several generations can be
produced in one year. So it facilitates rapid genetic analysis. The genome has
low repetitive DNA, over 60% of the nuclear DNA have protein coding functions.
5. The plant is small, self
fertilizes, annual long-day plant with short-life cycle (only 6 weeks), large
numbers of seeds are produced and they are easy to be grown in laboratory. It
is easy to induce mutations. It has many genomic resources and the
transformation can be done easily.
6. In 1982, Arabidopsis has
successfully completed its life cycle in Microgravity i.e. space. This shows
that Human Space Missions with plant companions may be possible.
Related Topics