Chapter: Biotechnology Applying the Genetic Revolution: Proteomics
Protein Tagging Systems
PROTEIN
TAGGING SYSTEMS
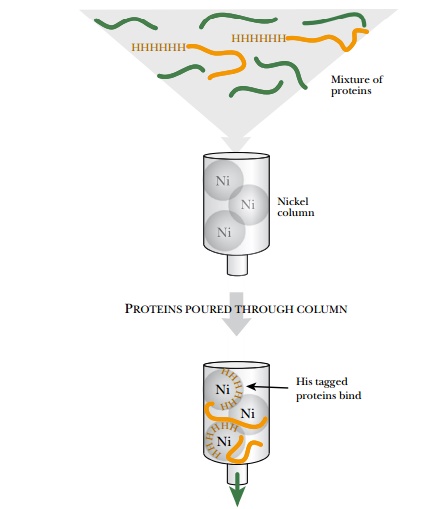
Protein tagging systems are
tools for the isolation and purification of single target proteins from a
mixture. The target protein is genetically fused to a segment of DNA that codes
for a “tag,” creating a hybrid gene. This is inserted into a vector with the
appropriate promoters and terminators to express the tagged protein of
interest. The gene construct is transformed into a suitable host organism for
expression. When the cells are grown and disrupted to release the proteins, the
target protein can be easily isolated because of its tag.
Many different types of tags
are used to isolate proteins because the chemistry and size of the tag may
affect the protein of interest in a negative way. The first widely used tag,
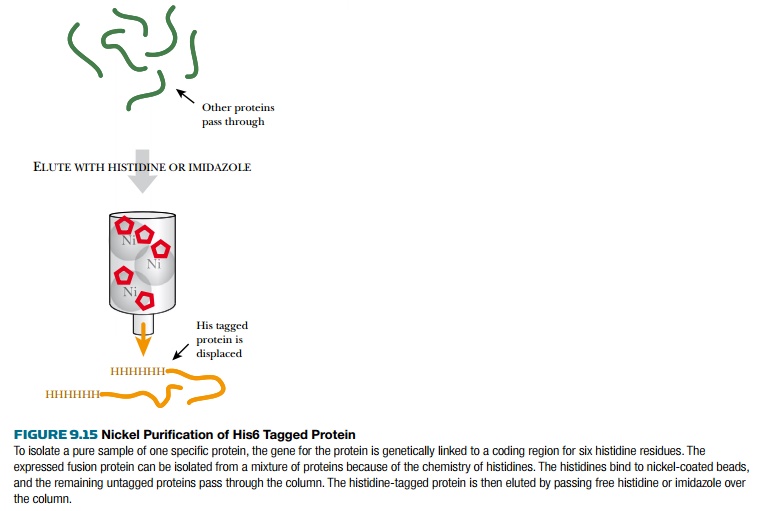
called the polyhistidine or His6 tag, consists of six histidine residues in a
row (Fig. 9.15). Histidine binds very tightly to nickel ions; therefore, His-tagged
proteins are purified on a column to which Ni2+ ions are attached. Once
attached to the column, the His6-tagged protein is removed by disrupting the
Ni2+–His interaction with free histidine or imidazole. The polyhistidine tag
may be attached to the carboxy- or amino-terminal end of the protein of
interest. Because the His6 tag is very short, the target protein is rarely
affected by adding it.
Other short tags for proteins
include FLAG, which is recognized by a specific antibody. FLAG has the peptide
sequence AspTyrLysAspAspAspAspLys. As before, the gene for the target protein
plus a short DNA segment encoding FLAG is cloned into a vector, and the hybrid
protein is produced in either bacteria or a cell line. FLAG-tagged proteins can
be isolated from a cell lysate using the anti-FLAG antibody either bound to
beads or attached to a column. Only the tagged protein attaches to the
beads/column. Finally, the FLAG-tagged protein is separated from the antibody
by adding free FLAG peptide. The short peptide is present in surplus and
competes for antibody with the tagged protein, which is therefore eluted from
the beads or column.
Another short tag is the
“Strep” tag, provided by a short DNA segment that encodes a 10-amino-acid
peptide with a similar structure to biotin. The biotin-like peptide binds
tightly to avidin and streptavidin, so Strep-tagged proteins are isolated by
binding to streptavidin-coated beads or a streptavidin column.
Besides short tags, longer tags that consist of entire proteins are used for some applications. Three popular tags include protein A from Staphylococcus, glutathione-S-transferase (GST) from Schistosoma japonicum, and maltose-binding protein (MBP) from Escherichia coli. Just like the short tags, the genes for these longer tags are genetically fused to the target gene. The hybrid gene constructs are expressed by using appropriate transcriptional promoters and terminators. Once the host cells express the hybrid gene, the fusion protein is isolated by purifying the protein tag. A specific antibody is available to bind to protein A, and lowering the pH releases the fusion protein. MBP binds to maltose (attached to beads or a column), and the fusion protein is released by adding free maltose. GST binds to its substrate, glutathione (on beads or a column), and free glutathione is used to release the hybrid protein. Once the fusion protein has been isolated, it must be cleaved to separate the target protein from the tag protein.
A useful feature of the
longer tags is the presence of protease cleavage site between the target
protein and the tagging protein.
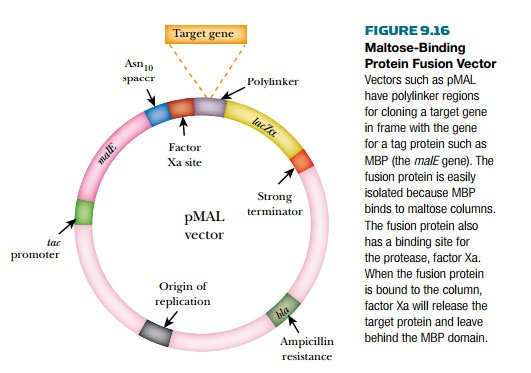
The vector for pMAL (New
England Biolabs, Inc., Ipswich, MA) has the gene for MBP, followed by a spacer
region, a recognition site for factor Xa, then the polylinker region for
cloning the target gene (Fig. 9.16). Factor Xa is a specific protease used in
the blood clotting system, and inserting its recognition sequence allows the
MBP portion of the hybrid protein to be cleaved from the target protein. After
the hybrid protein is eluted from the purification column with maltose, the
original protein is isolated by protease treatment. This is extremely useful when pure, native protein is needed
for analysis.
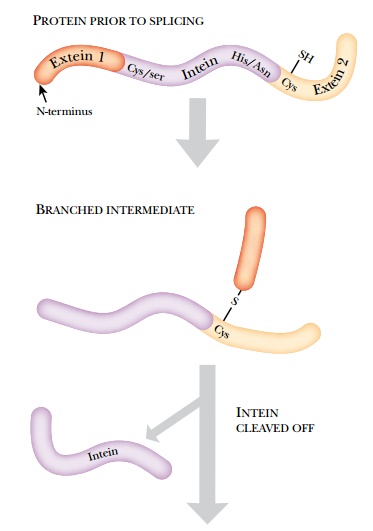
An even easier way to obtain
a pure sample of native protein is the self-cleavable intein tag. The approach
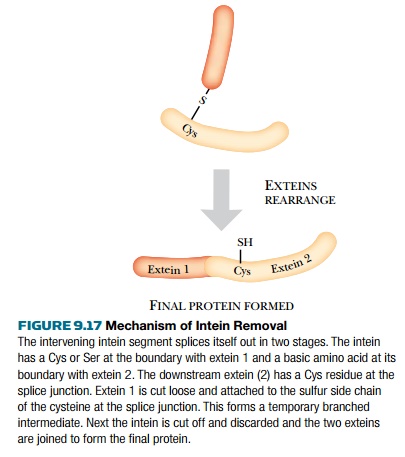
is based on inteins, self-splicing intervening segments found in some proteins.
Inteins are the protein equivalent to introns in RNA. The intein removes itself
from its host protein via a branched intermediate (Fig. 9.17).
The Intein Mediated
Purification with Affinity Chitin-binding Tag (IMPACT) system from New England
Biolabs uses a modified intein from the VMA1 gene of Saccharomyces cerevisiae.
Intein cleavage is used to release the target protein after purification of the
fusion protein (Fig. 9.18).
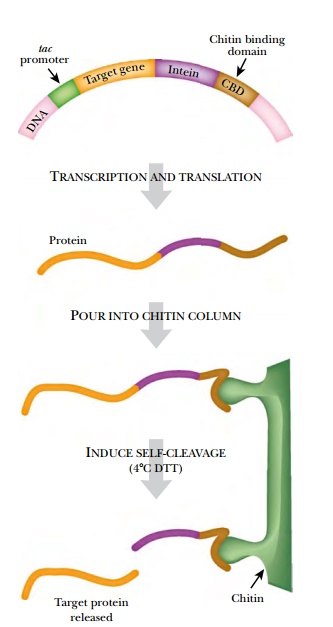

This yeast intein originally
cleaved both its N-terminus and its C-terminus, but it has been modified so
that it only cleaves its N-terminus. The chitin-binding tag of this system is
the small chitin-binding domain (CBD) from the chitinase A1 gene of Bacillus
circulans. (Chitin is the substance that
forms the exoskeleton of insects.) The vector has a polylinker or cloning site
for the target gene followed by the DNA segment encoding the intein, followed
by the CBD. The fusion protein is
expressed, and cell lysates containing the hybrid protein are isolated. When
the lysate passes through a chitin column, the hybrid protein binds to the
column via the CBD, and the remaining cellular proteins elute. The column is
then incubated at 4°C with dithiothreitol (DTT), a thiol reagent that activates
the intein to cleave its N-terminus. Thus the target protein is released from
the column, leaving behind the intein and CBD regions.
Related Topics