Chapter: Biotechnology Applying the Genetic Revolution: Proteomics
Mass Spectrometry for Protein Identification
MASS
SPECTROMETRY FOR PROTEIN IDENTIFICATION
Mass spectrometry is a
technique to determine the mass of molecules. In mass spectrometry, a molecule
is fragmented into different ions whose masses are accurately measured. The
ions generate a spectrum of peaks that is unique and therefore determines the
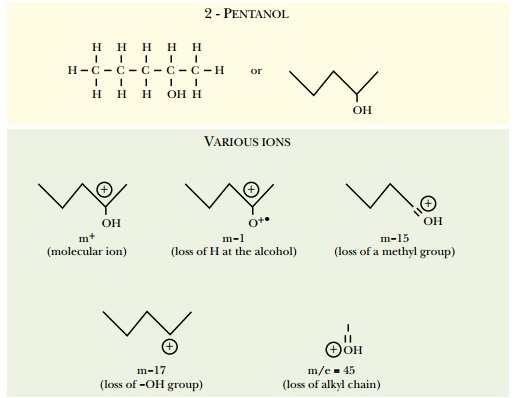
identity of the original molecule. The molecule 2-pentanol is used as an
example in Fig. 9.8. Here an electron beam aimed at the sample fragments the
2-pentanol into different ions: the molecular ion gains an electron, m-1 loses
hydrogen from the alcohol group, m-15 loses a methyl group, m-17 loses the
alcohol group, m/e = 45 loses the alkyl chain.
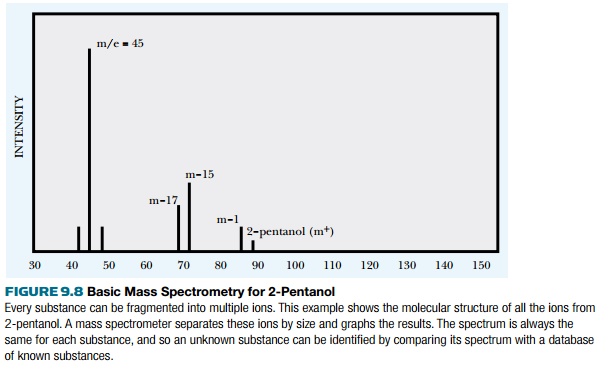
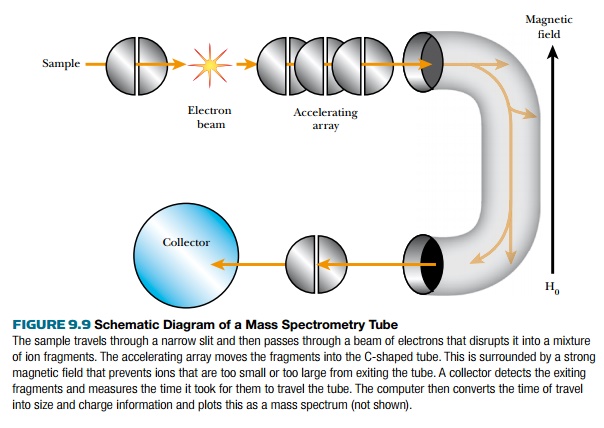
These ions are accelerated into a vacuum tube by an ion accelerating array. The ions travel through the tube at different speeds due to a magnetic field that causes the ions to follow a curved path within the tube (Fig. 9.9). The curves eliminate ions that are too small or too big. Ions that are too small gain so much momentum from the magnetic field that they collide with the wall. Those that are too big are not deflected by the magnetic field and also collide with the wall. Ions in the right size range are deflected by the magnetic field around both curves to hit the collector, where they are recorded as peaks in the mass spectrum. The base peak is the most intense, and other peaks are measured relative to this. The time ions take from the accelerator to the collector correlates directly to the size of the ion. Each peak is plotted based on the mass/charge ratio (m/z). The losses of mass (such as m-17 or m-15) refer to the loss of specific groups from the parent molecule and are most informative to the structure of the sample molecule because each such group has a characteristic mass.
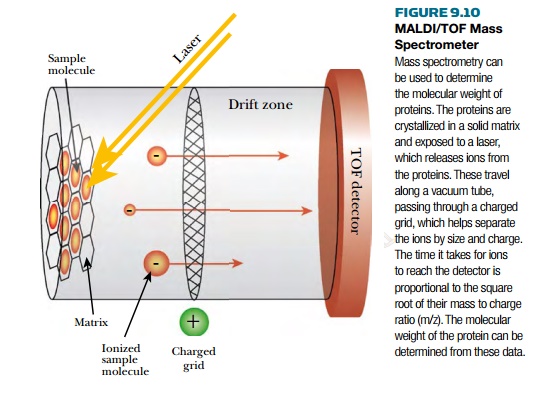
Until recently, very large
molecules such as proteins were beyond the range of mass spectrometry. Two
different ionization techniques have been developed that have made proteins
manageable. The first technique embeds proteins in a solid matrix before
ionization, and is called MALDI, or matrix-assisted laser desorption-ionization
(Fig. 9.10). Here the proteins are embedded in a material such as 4-methoxycinnamic
acid that absorbs laser light. The matrix absorbs and transfers the laser
energy to the proteins, causing them to release different ions. The ions are
accelerated through a vacuum tube by a charged grid. At the far end, the time-of-flight (TOF) detector records
the intensity and calculates the mass. In between is a flight tube that is free of electric fields. The ions are
accelerated with the same kinetic energy, and when they reach the flight tube,
the lighter ions move faster than the heavier ions. The time-of-flight is
proportional to the square root of mass to charge ratio (m/z). MALDI is able to
handle ions up to 100,000 daltons.
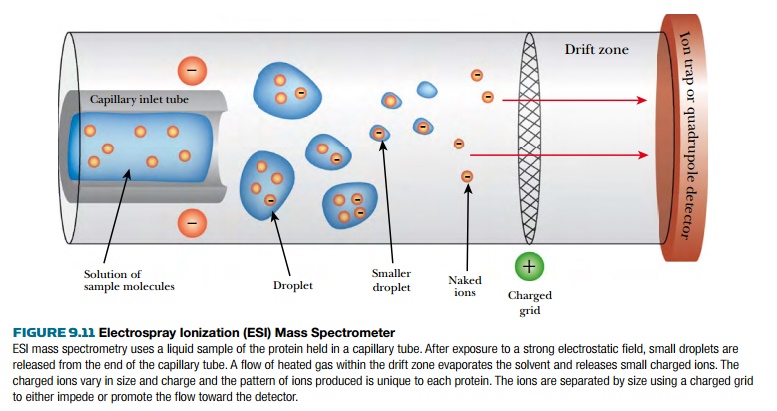
The other method for
preparing ions from proteins is electrospray
ionization (ESI; Fig. 9.11). Here the protein is dissolved in liquid and very small droplets are released from a
narrow capillary tube. The droplets enter the electrostatic field, where a
heated gas, such as hydrogen, causes the solvent to evaporate and the droplets
to break up. This causes the protein to release ions into the vacuum tube,
where they are accelerated by the electric field. The detector at the far end
varies based on the sample being studied.
A TOF detector may be used,
as described earlier. Other detectors use quadrupole ion traps or Fourier
transform ion cyclotron resonance to determine the mass of the ions.
Quadrupole ion traps capture
the ions in an electric field.
The ions are then ejected into the detector by a second electric field. The electric field controls what size ions can pass to the detector, and varying the field allows different-sized ions to be detected. Combination detectors exist that use both TOF and quadrupole ion traps. The advantage that ESI has over MALDI is that proteins isolated from HPLC (see earlier discussion) require no special preparation and can be used directly. The disadvantage of ESI is that masses of about 5000 are the maximum.
The use of mass spectroscopy
will become more prevalent as techniques improve. For example, surface-enhanced
laser desorption-ionization (SELDI) mass spectroscopy takes liquid samples and
ionizes the proteins that adhere to a treated metal bar. The technique shows
great promise for bodily fluids such as blood and, it is hoped, will help in
identifying a particular protein profile for different disease states. Perhaps
one day patients will be diagnosed with cancer long before any symptoms are
detected. A change in the protein profile in their blood could denote that
cancer cells are forming. MALDI and ESI are sensitive enough to detect changes
in proteins due to phosphorylation, glycosylation, and so forth. The technique
can identify which amino acid is modified because only one specific ion is
altered.
Related Topics