Chapter: Biotechnology Applying the Genetic Revolution: Proteomics
Phage Display Library Screening
PHAGE
DISPLAY LIBRARY SCREENING
A phage display library is a collection of bacteriophage particles
that have segments of foreign proteins protruding from their surface (Fig.
9.19). Normal bacteriophages have outer coats made of proteins. The outer coat
of M13 bacteriophage has about 2500 copies of the major coat protein (gene VIII
protein) and about five copies of the minor coat protein (gene III protein).
One particular phage display system fuses gene III protein to the foreign
protein, so that M13 now has about five copies of the foreign protein on its
surface. M13 is convenient because it does not lyse the bacteria it infects;
the viral particles are simply secreted through the bacterial cell envelope.
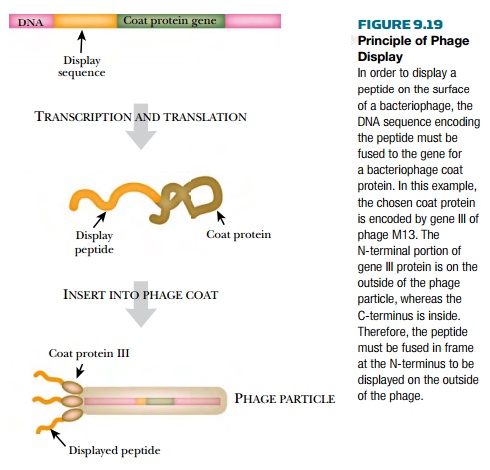
In order for the phage to
display a foreign protein, the gene for that protein must be fused to gene III
to produce a hybrid protein. The gene of interest must be in frame with gene
III for proper expression. The M13 genome can accommodate extra DNA because the
bacteriophage particles are filamentous and are made longer to package a larger
genome. The M13 genome containing the gene of interest is transformed into E. coli, where the bacteriophage DNA
directs the synthesis of new
particles containing the protein of interest in the coat.
Bacteriophage can also
display small segments of genes. Random oligonucleotides generated by PCR can
be cloned and fused to gene III of M13. Each random oligonucleotide will encode
a different peptide. These clones are transformed into the bacteriophage, and
each transformant will display its foreign peptide fused to gene III protein.
The collection of displayed
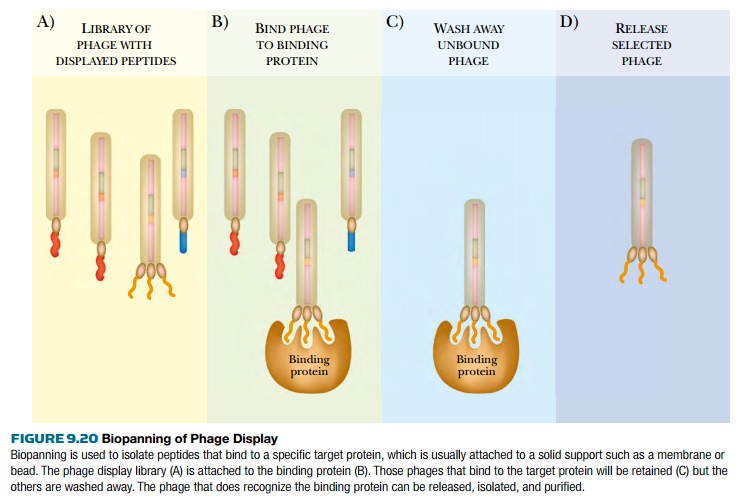
peptides can be useful in many ways. They can be screened by biopanning to find a particular target,
perhaps a specific protein binding domain, or a specific peptide that binds an
antibody (Fig. 9.20). In biopanning, the library of phages displaying the
foreign peptides is incubated with the target, such as an antibody, bound to a
bead or membrane. All the recombinant phages that bind to that antibody adhere
to the solid support, and the others are washed away. All the bound phages are
eluted and incubated with E. coli to
replicate the phage. The procedure is usually repeated in order to enrich for
peptides that bind specifically,
because some nonspecific binding could occur. Once a phage with a useful
peptide is identified, the clone is sequenced to determine the structure of the
peptide.
Full-length protein libraries can also be studied using phage display, but pose some extra problems. Coding sequences for full-length proteins must be cloned in frame with both the signal sequence at the N-terminal end and gene III at the C-terminal end. (The signal sequence is required to direct the hybrid protein to the viral coat.) Ensuring the correct reading frame is reasonable for one or two genes, but for an entire library, there is too much room for error. Besides, the possible creation of a stop codon at either fusion junction would prevent the hybrid protein from being expressed. The solution is to use T7 bacteriophage for libraries of full-length proteins. T7 has a coat protein whose C-terminal tail is exposed to the outside. To be expressed on the bacteriophage surface, the protein library must therefore be fused to the C-terminus of the coat protein. This requires only one fusion junction. Furthermore, even if library sequences are cloned out of frame or contain stop codons, the coat protein itself is unaffected and still assembled, although the attached library proteins will be defective.
Being able to express
full-length proteins for biopanning is very useful to proteomics researchers.
To identify a protein that binds to a particular cell surface receptor, a phage
display library can be biopanned for receptor binding. Another example is
finding RNA binding proteins. Here, RNA is anchored to a solid support and the
phage display library is incubated with this RNA “bait.” The phages that stick
to the RNA bait are isolated and enriched by repeating the procedure. Each
isolated clone can then be sequenced to identify which proteins bind RNA.
Related Topics