Chapter: Biotechnology Applying the Genetic Revolution: Proteomics
Protein Arrays
PROTEIN ARRAYS
Protein-detecting arrays may
be divided into those that use antibodies and those based on using tags. In the
ELISA assay, antibodies to specific proteins are attached to a solid support,
such as a microtiter plate or glass slide. The protein sample is then added and
if the target protein is present, it binds its complementary antibody. Bound
proteins are detected by adding a labeled second antibody.
Another antibody-based
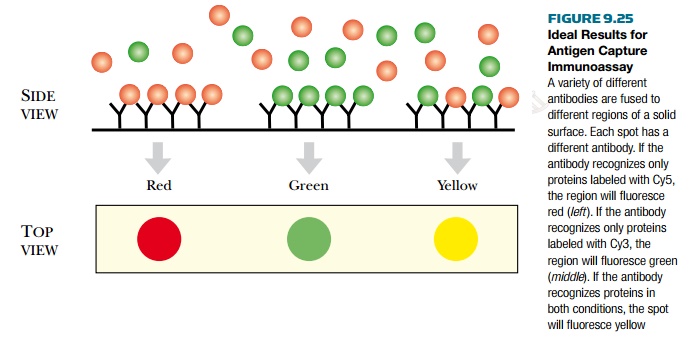
protein-detecting array is the antigen capture immunoassay (Fig. 9.25). Much
like the ELISA, this method uses antibodies to various proteins bound to a
solid surface. The experimental protein sample is isolated and labeled with a
fluorescent dye. If two conditions are being compared, proteins from sample 1
can be labeled with Cy3, which fluoresces green, and proteins from sample 2 can
be labeled with Cy5, which fluoresces red. The samples are added to the
antibody array, and complementary proteins bind to their cognate antibodies. If
both sample 1 and 2 have identical proteins that bind the same antibody, the
spot will fluoresce yellow. If sample 1 has a protein that is missing in sample
2, then the spot will be green. Conversely, if sample 2 has a protein missing
from sample 1, the spot will be red. This method is good for comparing protein
expression profiles for two different conditions.
In the third method, the
direct immunoassay or reverse-phase array, the proteins of the experimental
sample are bound to the solid support (Fig. 9.26). The proteins are then probed
with a specific labeled antibody.
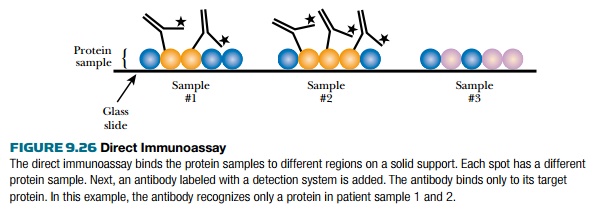
Both presence and amount of
protein can be monitored. For example, proteins from different patients with
prostate cancer can be isolated and spotted onto glass slides. Each sample can
be examined for specific protein markers or the presence of different cancer
proteins. The levels of certain proteins may be related to the stages of
prostate cancer. This immunoassay helps researchers to decipher these
correlations.
The main problem with
immuno-based arrays is the antibody. Many antibodies cross-react with other
cellular proteins, which generates false positives. In addition, binding
proteins to solid supports may not be truly representative of intracellular
conditions. The proteins are not purified or separated; therefore, samples contain
very diverse proteins. Some proteins will bind faster and better than others.
Also, proteins of low abundance may not compete for binding sites. Another problem is that many proteins are
found in complexes, so other proteins in the complex may mask the antibody
binding site.
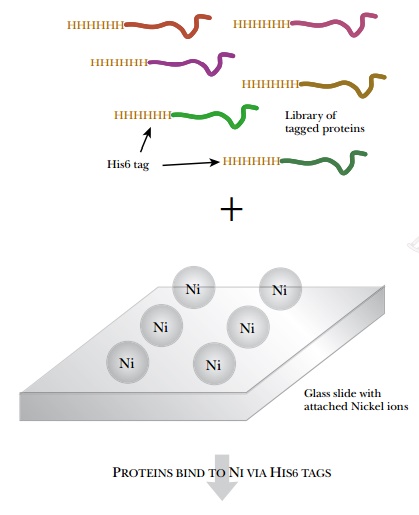
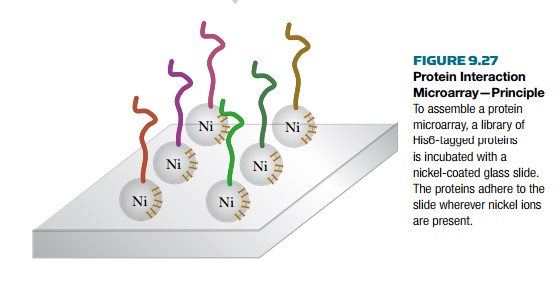
Rather than using antibodies,
protein interaction arrays use a fusion tag to bind the protein to a solid
support (Fig. 9.27). The use of protein arrays to determine protein
interactions and protein function is a natural extension of yeast two-hybrid
assays and co-immunoprecipitation. Protein arrays can assess thousands of
proteins at one time, making this a powerful technique for studying the
proteome. Protein arrays are often used in yeast because its proteome contains
only about 6000 proteins. Libraries have been constructed in which each protein
is fused to a His6 or GST tag. The proteins are then attached by the tags to a
solid support such as a glass slide coated with nickel or glutathione. To build
the array, each protein is isolated individually and spotted onto the glass
slide. The tagged proteins bind to the slide and other cellular components are
washed away. Each spot has only one unique tagged protein.
Once the array is assembled,
the proteins can be assessed for a particular function. In the laboratory of
Michael Snyder at Yale University, the yeast proteome has been screened for
proteins that bind calmodulin (a
small Ca2+ binding protein) or phospholipids (Fig. 9.28). Both
calmodulin and phospholipid were tagged with biotin and incubated with a slide
coated with each of the yeast proteins bound to the slide via
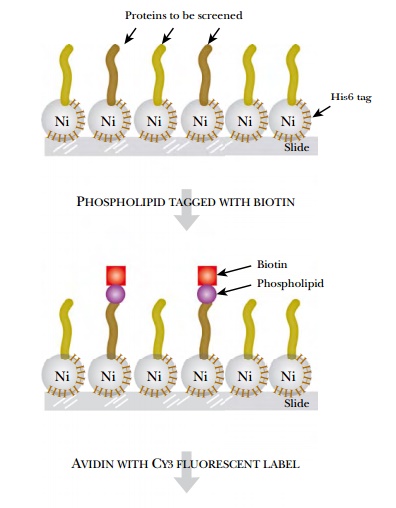
His6-nickel interactions. The
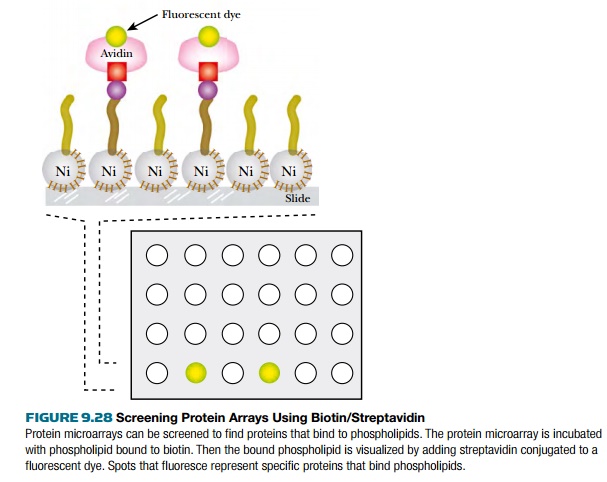
biotin-labeled calmodulin or phospholipid was then visualized by incubating the
slide with Cy3-labeled streptavidin. (Streptavidin binds very strongly to
biotin.) The results identified 39 different calmodulin binding proteins (only
six had been identified previously), and 150 different phospholipid binding
proteins.
Related Topics