Chapter: Biotechnology Applying the Genetic Revolution: DNA Synthesis in Vivo and in Vitro
PCR in Genetic Engineering
PCR IN GENETIC ENGINEERING
PCR allows the scientist to
clone genes or segments of genes for identification and analysis. PCR also
allows the scientist to manipulate a gene that has already been identified.
Various modifiedPCR techniques allow scientists to hybridize two separate genes
or genes segments into one, delete or invert regions of DNA, and alter single
nucleotides to change the gene and its encoded protein in a more subtle way.
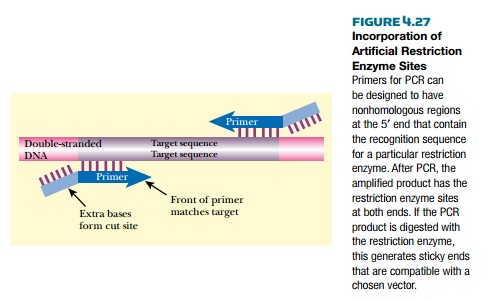
PCR can make cloning a foreign piece of DNA easier. Special PCR primers can generate new restriction enzyme sites at the ends of the target sequence (Fig. 4.27). The primer is synthesized so that its 5′ end has the desired restriction enzyme site, and the 3′ end has sequence complementary to the target. Obviously, the 5′ end of the primer does not bind to the target DNA, but as long as the 3′ end has enough matches to the target, then the primer will still anneal. Taq polymerase primes synthesis from the 3′ end; therefore, the enzyme is not bothered by mismatched 5′ sequences. The resulting PCR product can easily be digested with the corresponding restriction enzyme and ligated into the appropriate vector.
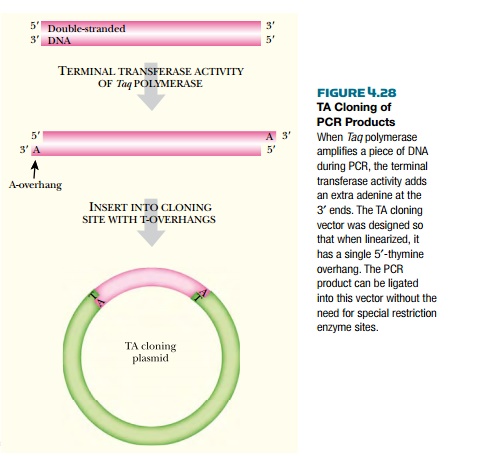
Rather than incorporating
restriction enzyme sites into the ends of the PCR product, TA cloning will
clone any PCR product directly (Fig. 4.28). Taq polymerase has terminal
transferase activity that generates a single adenine overhang on the ends of
the PCR products it makes. Special vectors containing a single thymine overhang
have been developed, and simply mixing the PCR product with the TA cloning
vector plus DNA ligase clones the PCR product into the vector without any
special modifications.
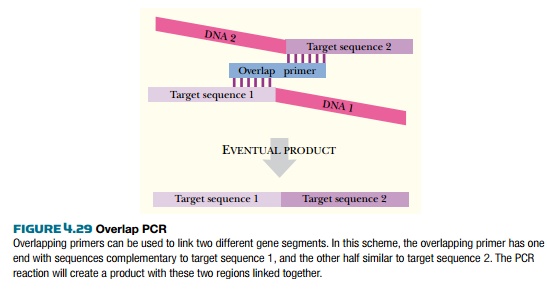
PCR can be used to manipulate
cloned genes also. Two different gene segments can be hybridized into one using
overlap PCR (Fig. 4.29). Here, PCR amplification occurs with three primers: one
is complementary to the beginning of the first gene segment, one is
complementary to the end of the second gene segment, and a third is half
complementary to the end of gene segment 1 and half complementary to the
beginning of gene segment 2. During PCR, the two gene segments become fused
into one by a mechanism that is hard to visualize, but probably involves
looping of some of the early PCR products.
PCR can be used to create large deletions or insertions into a gene (Fig. 4.30). Once again the design of the PCR primers is key to the construction. For example, primers to generate insertions have two regions: the first half is homologous to the sequence around the insertion point; the second half has sequences complementary to the insert sequence. For example, suppose an antibiotic resistance gene such as npt (confers resistance to neomycin) is to be inserted into a cloning vector. The primers would have their 5′ ends complementary to the sequence flanking the insertion point on the vector and their 3′ ends complementary to the ends of the npt gene. First the primers are used to amplify the npt gene and give a product with sequences homologous to the vector flanking both ends. Next, the PCR product is transformed into bacteria harboring the vector. The npt gene recombines with the insertion point by homologous recombination, resulting in insertion of the npt gene into the vector.
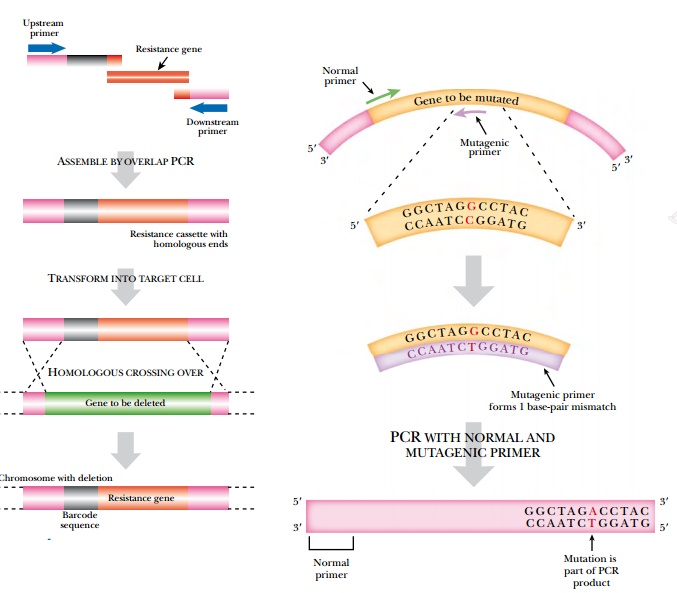
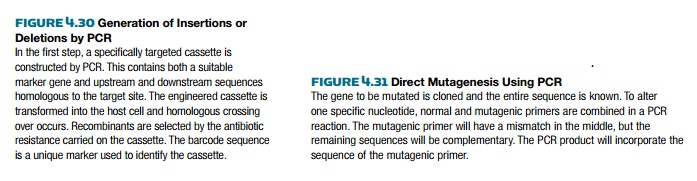
The insertion point(s) will
determine whether the antibiotic cassette causes just an insertion or both an
insertion plus a deletion. If the two PCR primers recognize separate homologous
recombination sites, then the incoming PCR segment will recombine at these two
sites.
Homologous recombination then
results in the npt gene replacing a
piece of the vector rather than merely inserting at one particular location.
PCR can also generate nucleotide changes in a gene by directed mutagenesis (Fig. 4.31). Usually only one or a few adjacent nucleotides are changed. First, a mutagenic PCR primer is synthesized that has nucleotide mismatches in the middle region of the primer. The primer will anneal to the target site with the mismatch in the center. The primer needs to have enough matching nucleotides on both sides of the mismatch so that binding is stable during the PCR reaction. The mutagenic primer is paired with a normal primer. The PCR reaction then amplifies the target DNA incorporating the changes at the end with the mutagenic primer. These changes may be relatively subtle, but if the right nucleotides are changed, then a critical amino acid may be changed. One amino acid change can alter the entire function of a protein. Such an approach is often used to assess the importance of particular amino acids within a protein.
Related Topics