Chapter: Biotechnology Applying the Genetic Revolution: DNA Synthesis in Vivo and in Vitro
Modifications of Basic PCR
MODIFICATIONS
OF BASIC PCR
Many different permutations
of PCR have been devised since Kary Mullis developed the basic procedure. All
rely on the same basic PCR reaction, which takes a small amount of DNA and
amplifies it by in vitro replication. Many of these variant protocols are
essential tools for recombinant DNA research.
Several strategies allow
amplifying a DNA segment by PCR even if its
sequence is unknown. For example, the unknown sequence may be cloned
into a vector (whose sequence is known). The primers are then designed to anneal
to the regions of the vector just outside the insert.
In another scenario, the
sequence of an encoded protein is used to generate PCR primers. Remember that
most amino acids are encoded by more than one codon. Thus, during translation
of a gene, one or more codons are used for the same amino acid. Therefore, if a
protein sequence is converted backwards into nucleotide sequence, the sequence
is not unique. For example, two different codons exist for histidine and
glutamine, and four codons exist for serine. Consequently, the nucleotide
sequence encoding the amino acid sequence histidine-glutamine-valine can be one
of 16 different combinations (Fig. 4.23).
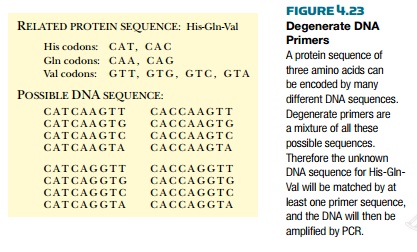
If primers are made that
depend on protein sequence they will be degenerate primers and they will have a mixture of two or three
different bases at the wobble positions in the triplet codon.
During oligonucleotide
synthesis, more than one phosphoramidite nucleotide can be added to the column
at a particular step. Some of the primers will have one of the nucleotides, whereas other primers will have
the other nucleotide. If many different wobble bases are added, a population of
primers are created, each with a slightly different sequence.
Within this population, some
will bind to the target DNA perfectly, some will bind with only a few mismatches, and some won’t bind
at all. Of course, the annealing temperature for degenerate primers is adjusted
to allow for some mismatches.
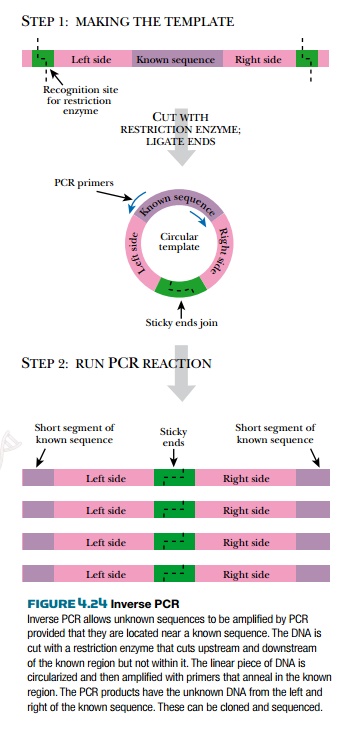
Inverse PCR is a trick used when sequence information is known only on one side of the target region (Fig. 4.24). First, a restriction enzyme is chosen that does not cut within the stretch of known DNA. The length of the recognition sequence should be six or more base pairs in order to generate reasonably long DNA segments for amplification by PCR. The target DNA is then cut with this restriction enzyme to yield a piece of DNA that has compatible sticky ends, one upstream of the known sequence, and one downstream. The two ends are ligated to form a circle. The PCR primers are designed to recognize the end regions of the known sequence. Each primer binds to a different strand of the circular DNA and they both point “outwards” into the unknown DNA. PCR then amplifies the unknown DNA to give linear molecules with short stretches of known DNA at the ends, and the restriction enzyme site in the middle.
Related Topics