Chapter: Biotechnology Applying the Genetic Revolution: DNA Synthesis in Vivo and in Vitro
Automated DNA Cycle Sequencing Combines PCR and Sequencing
AUTOMATED
DNA CYCLE SEQUENCING COMBINES PCR AND SEQUENCING
Automated DNA sequencing uses
a PCR-type reaction to sequence DNA. This is an improved way to sequence DNA
because of its speed and because it can be analyzed by computer rather than a
person. In PCR sequencing, or cycle
sequencing, the template DNA with unknown sequence is amplified by Taq polymerase rather than Klenow
polymerase. As for DNA polymerase I, this Taq
DNA polymerase was modified to remove its proofreading ability. Cycle
sequencing reaction mixtures includes all four deoxynucleotides, all four
dideoxynucleotides, a single primer, template DNA, and Taq polymerase. Unlike regular sequencing, each dideoxynucleotide
is labeled with a fluorescent tag of a different color; therefore, all four
dideoxynucleotides are in the same reaction mix.
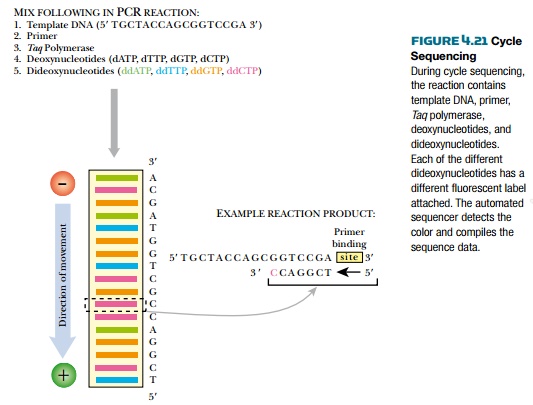
The samples are amplified in
a thermocycler. First, the template DNA is denatured at a high temperature,
then the temperature is lowered to anneal the primer, finally the temperature
is raised to 70°C, the optimal temperature for Taq polymerase to make DNA copies of the template. During
polymerization, dideoxynucleotides are incorporated and cause chain
termination, just as with standard sequencing. The ratio of dideoxynucleotides
to deoxynucleotides is adjusted to ensure that some fragments stop at each G,
A, T, or C. After Taq polymerase makes thousands of copies of the template,
each stopping at a different nucleotide, the entire mixture is run in one lane
of a sequencing gel (Fig. 4.21). Bands of four different colors are seen,
corresponding to the four fluorescently labeled dideoxynucleotides and hence
the four bases.
Cycle sequencing has some advantage over regular sequencing. During regular sequencing reactions, the temperatures are lower; therefore, the template DNA is more likely to fold back on itself into different secondary structures. This is especially true for double-stranded DNA templates, which can reanneal before Klenow polymerase has a chance to work. During cycle sequencing, each round brings the temperature to 95°C, which destroys any secondary structures or double-stranded regions. Another advantage of cycle sequencing is to control primer hybridization. Some primers do not work well with regular sequencing reactions because they bind to closely related sequences. During cycle sequencing, the primer annealing temperature is controlled and can be set quite high in order to combat nonspecific binding. Finally, cycle sequencing requires much less template DNA than regular sequencing; therefore, sequencing can be done from smaller samples.
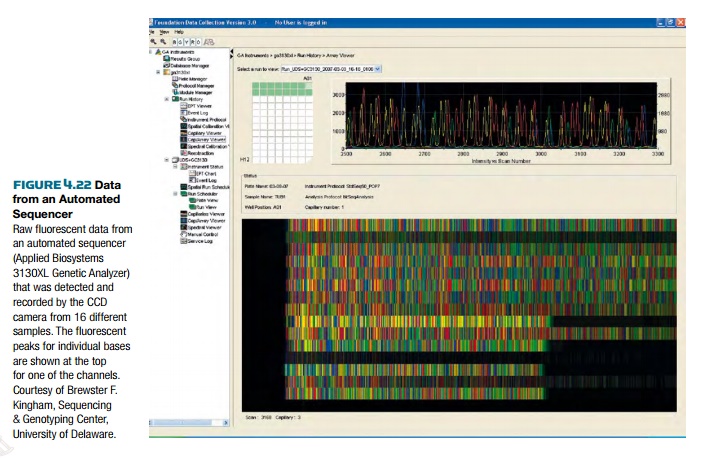
Another advance in sequencing
has been the detection system. Traditionally, researchers have used
radioactively labeled nucleotides, followed by autoradiography to detect the
bands. A major improvement has been to use fluorescently labeled
dideoxynucleotides; therefore, the terminal nucleotide of each DNA strand will
fluoresce. Different fluorescent tags can be used for each of the four
dideoxynucleotides, so that the whole reaction can be done in one tube. Automatic DNA sequencers detect each of
the fluorescent tags and record the sequence of bases (Fig. 4.22). Much like regular sequencing, the different
fragments are run on a gel, but instead of four lanes, the entire mix is all in
the same lane. Some automatic DNA sequencers can read up to 96 samples
side-by-side on each gel. At the bottom of each lane is a fluorescent
activator, which emits light to excite the fluorescent dyes. On the other side
is the detector, which reads the wavelength of light that the fluorescent dye
emits. As each fragment passes the detector, it measures the wavelength and
records the data as a peak on a graph. For each fluorescent dye, a peak is
recorded and assigned to the appropriate base. An attached computer records and
compiles the data into the DNA sequence.
Automated sequencing has a
large startup cost because the sequence analyzer is quite expensive, but they
run multiple samples at one time, and thus the cost per sample is quite low.
Many universities and companies have a centralized facility that does the
sequencing for all the researchers. In fact, sequencing has become so automated
that many researchers just send their template DNA and primers to a company
that specializes in sequencing.
Related Topics