Chapter: 11th Botany : Chapter 5 : Taxonomy and Systematic Botany
Modern trends in taxonomy
Modern trends in taxonomy
Taxonomists now accept that, the morphological
characters alone should not be considered in systematic classification of
plants. The complete knowledge of taxonomy is possible with the principles of
various disciplines like Cytology, Genetics, Anatomy, Physiology, Geographical
Distribution, Embryology, Ecology, Palynology, Phenology, Bio-Chemistry,
Numerical Taxonomy and Transplant Experiments. These have been found to be
useful in solving some of the taxonomical problems by providing additional
characters. It has changed the face of classification from alpha (classical) to omega
(modern kind). Thus the new systematic has evolved into a better taxonomy.
1. Chemotaxonomy
Various medicines, spices and preservatives
obtained from plant have drawn the attention of Taxonomists. Study of various
chemicals available in plants help to solve certain taxonomical problems.
Chemotaxonomy is the scientific approach of classification of plants on the
basis of their biochemical constituents. As proteins are more closely
controlled by genes and less subjected to natural selection, it has been used
at all hierarchical levels of classification starting from the rank of
ŌĆśvarietyŌĆÖ up to the rank of division in plants. Proteins, amino acids, nucleic
acids, peptides etc. are the most studied chemicals in chemotaxonomy.
The chemical characters can be divided into three main categories.
1. Easily visible characters like starch grains, silica etc.
2. Characters detected by chemical tests like phenolics, oil, fats, waxes etc.
3. Proteins
Aims of chemotaxonomy
1.
To develop taxonomic characters which may improve
existing system of plant classification.
2.
To improve present day knowledge of phylogeny of
plants.
2. Biosystematics
Biosystematics is an ŌĆ£Experimental, ecological and
cytotaxonomyŌĆØ through which life forms are studied and their relationships are
defined. The term biosystematics was introduced by Camp and Gilly in 1943.
Many authors feel Biosystematics is closer to Cytogenetics and Ecology and much
importance given not to classification but to evolution.
Aims of biosystematics
The aims of biosystematics are as follows:
1.
To delimit the naturally occurring biotic community
of plant species.
2.
To establish the evolution of a group of taxa by
understanding the evolutionary and phylogenetic trends.
3.
To involve any type of data gathering based on
modern concepts and not only on morphology and anatomy.
4.
To recognize the various groups as separate
biosystematic categories such as ecotypes, ecospecies, cenospecies and
comparium.
3. Karyotaxonomy
Chromosomes are the carriers of genetic
information. Increased knowledge about the chromosomes have been used for
extensive biosystematic studies and resolving many taxonomic problems.
Utilization of the characters and phenomena of cytology for the explanation of
taxonomic problem is known as cytotaxonomy
or karyotaxonomy. The characters of chromosome such as
number, size, morphology and behaviour during meiosis have proved to be of taxonomic
value.
4. Serotaxonomy (immunotaxonomy)
Systematic
serology or serotaxonomy had its
origin towards the end of twentieth century with the discovery of serological
reactions and development of the discipline of immunology. The classification
of very similar plants by means of differences in the proteins they contain, to
solve taxonomic problems is called serotaxonomy.
Smith (1976) defined it as ŌĆ£the study of the origins and properties of
antisera.ŌĆØ
Importance of serotaxonomy
It determines the degree of similarity between
species, genera, families etc. by comparing the reactions of antigens from
various plant taxa with antibodies raised against the antigen of a given taxon.
Example: 1. The assignment of Phaseolus aureus and P. mungo
to the genus Vigna is strongly
supported by serological evidence by Chrispeels and Gartner.
5. Molecular taxonomy (molecular systematics / molecular phylogenetics)
Molecular Taxonomy is the branch of phylogeny that
analyses hereditary molecular differences, mainly in DNA sequences, to gain
information and to establish genetic relationship between the members of
different taxonomic categories. The advent of DNA cloning and sequencing
methods have contributed immensely to the development of molecular taxonomy and population genetics over the years. These
modern methods have revolutionised the field of molecular taxonomy and
population genetics with improved analytical power and precision.
Molecular Markers
Allozyme electrophoresis is a method which
can identify genetic variation at the level
of enzymes that are directly encoded by DNA.
Mitochondrial DNA markers are non- nuclear
DNA located within organelles in the
cytoplasm called mitochondria. The entire genome undergoes transcription as one
single unit. They are not subjected to recombination and thus they are
homologous markers.
Microsatellite is a simple DNA
sequence which is repeated several times across various points in the DNA of an organism. These (usually 2-5)
repeats are highly variable and these loci can be used as markers. (Example:
TGTGTG, in which two base pairs repeat, the region are termed tandem repeat.)
Single nucleotide polymorphisms arise due to single nucleotide substitutions (transitions/transversions) or single nucleotide
insertions/deletions. SNPs are the most abundant polymorphisms in the genome
(coding and non-coding) of any organism. These single nucleotide variants can
be detected using PCR, microchip arrays or fluorescence technology.
DNA microarray or DNA chip consists of small
glass microscope slides, silicon chip
or nylon membranes with many immobilized DNA fragments arranged in a standard
pattern. A DNA microarray can be utilized as a medium for matching a reporter
probe of known sequence against the DNA isolated from the target sample which
is of unknown origin. Species-specific DNA sequences could be incorporated to a
DNA microarray and this could be used for identification purposes.
Arbitrary markers are sometimes used
to target a segment of DNA of unknown function.
The widely used methods of amplifying unknown regions are RAPD and AFLP DNA.
Specific Nuclear DNA markers: Variable Number of
Tandem Repeat is a segment of DNA
that is repeated tens or even hundreds to thousands of times in nuclear genome.
They repeat in tandem; vary in number in different loci and differently in
individuals. There are two main classes of repetitive and highly polymorphic
DNA viz. minisatellite DNA referring
to genetic loci with repeats of length 9-65 bp and microsatellite DNA with
repeats of 2-8 bp (1-6) long. Microsatellites are much more numerous in the
genome of vertebrates than minisatellites.
![]()
![]()
![]()
The results of a molecular phylogenetic analysis
are expressed in the form of a tree called phylogenetic tree. Different molecular markers like allozymes, mitochondrial DNA, micro satellites, RFLP
(Restriction Fragment Length Polymorphism), RAPD (Random amplified polymorphic
DNA), AFLPs (Amplified Fragment Length Polymorphism), single nucleotide
polymorphism- SNP, microchips or arrays are used in analysis.
Uses of molecular taxonomy
1. Molecular taxonomy helps in establishing the
relationship of different plant groups at DNA level.
2.
It unlocks the treasure chest of information on
evolutionary history of organisms.
RFLP (Restriction Fragment Length Polymorphism)
RFLPs is a
molecular method of genetic analysis
that allows identification of taxa based on unique patterns of restriction
sites in specific regions of DNA. It refers to differences between taxa in
restriction sites and therefore the lengths of fragments of DNA following
cleavage with restriction enzymes.
Amplified Fragment Length Polymorphism (AFLP)
This method is similar to that of identifying RFLPs
in that a restriction enzyme is used to cut DNA into numerous smaller pieces,
each of which terminates in a characteristic nucleotide sequence due to the
action of restriction enzymes.
AFLP is
largely used for population genetics
studies, but has been used in studies of closely related species and even in
some cases, for higher level cladistic analysis.
Random Amplified Polymorphic DNA (RAPD)
It is a method to identify genetic markers using a
randomly synthesized primer that will anneal (recombine (DNA) in the double
stranded form) to complementary regions located in various locations of isolated
DNA. If another complementary site is present on the opposing DNA strand at a
distance that is not too great (within the limits of PCR) then the reaction
will amplify this region of DNA.
RAPDs like microsatellites may often be used for
genetic studies within species but may also be successfully employed in
phylogenetic studies to address relationships within a species or between
closely related species. However RAPD analysis has the major disadvantage that
results are difficult to replicate and in that the homology of similar bands in
different taxa may be nuclear.
Significance of Molecular Taxonomy
1.
It helps to identify a very large number of species
of plants and animals by the use of conserved molecular sequences.
2.
Using DNA data evolutionary patterns of
biodiversity are now investigated.
3.
DNA taxonomy plays a vital role in phytogeography,
which ultimately helps in genome mapping and biodiversity conservation.
4.
DNA- based molecular markers used for designing DNA
based molecular probes, have also been developed under the branch of molecular
systematics.
6. DNA Barcoding
Have you seen how scanners are used in supermarkets
to distinguish the Universal Product Code (UPC)? In the same way we can also distinguish one species
from another. DNA barcoding is a taxonomic method that uses a very short
genetic sequence from a standard part of a genome. The genetic sequence used to
identify a plant is known as ŌĆ£DNA tagsŌĆØ
or ŌĆ£DNA barcodesŌĆØ. Paul Hebert in 2003 proposed ŌĆśDNA
barcodingŌĆÖ and he is considered
as ŌĆśFather of barcodingŌĆÖ.
The gene region that is being used as an effective
barcode in plants is present in two genes of the chloroplast, matK and rbcL,
and have been approved as the barcode regions for plants.
Sequence of unknown species can be matched from
submitted sequence in GenBank using Blast (web-programme for searching the
closely related sequence).
Significance of DNA barcoding
1.
DNA barcoding greatly helps in identification and
classification of organism.
2.
It aids in mapping the extent of biodiversity.
DNA barcoding techniques require a large database
of sequences for comparison and prior knowledge of the barcoding region.
However, DNA barcoding is a helpful tool to
determine the authenticity of botanical material in whole, cut or powdered
form.
List of conferences of International Barcode was
held
S.No. Year Place
1.
2005
London, United Kingdom
2.
2007
Taipei, Taiwan
3.
2009
Mexico City, Mexico
4.
2011
Adelaide, Australia
5.
2013
Yunnan, China
6.
2015
Guelph, Canada
7.
20-24 Skukuza, South Africa Nov' 2017
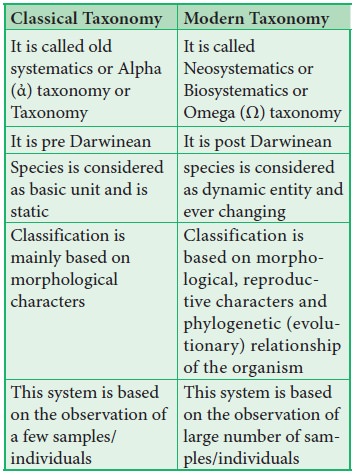
7. Differences between classical and modern taxonomy
Related Topics