Chapter: Biology of Disease: Transfusion and Transplantation
HLA System and Typing
THE HLA SYSTEM
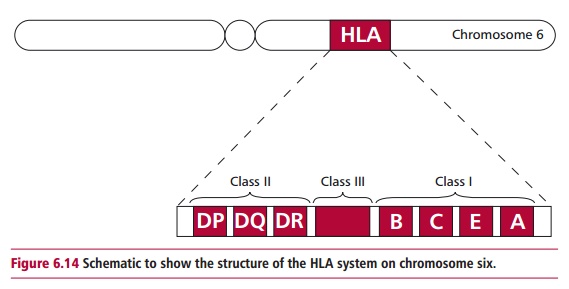
The HLA system of genes is found on the short arm of chromosome 6. This region encodes MHC proteins. The structures of MHC I and II were discussed. Here their involvement in triggering rejection will be emphasized. Molecules of MHC I consist of a single polypeptide encoded by the MHC that is always associated with a smaller protein, β 2M, encoded outside the MHC. However, MHC II pro-teins consist of two polypeptides, α and β, both of which are encoded by the MHC region. Figure 6.14illustrates the structure of the HLA-region although it has beengreatly simplified to aid understanding. The HLA complex contains a number of genetic loci, including those that encode different types of Class I proteins. Thus, the HLA-A, B and C regions contain genes that encode HLA-A, B and C proteins respectively. These are all found on nucleated cells and are distinct types of proteins and not allelic forms of each other. However, there are allelic forms of each of the HLA-A, B and C genes, and these encode HLA proteins, each of which have small differences in their amino acid sequences. Each of the Class I genes is polyallelic, which means that many different alleles exist although, of course, each individual only expresses a maximum of two alleles at each locus, given chromosomes occur in pairs Moreover, the alleles are codominant so that each nucleated cell expresses two different alle-les of the HLA-A gene, as well as two different forms of the HLA-B and of the HLA-C gene. A large number of allelic forms of each gene exist (Table 6.13).
The HLA complex in humans is one of the most highly polymorphic systems known. Given that each individual has two of each of these alleles, and that the allelic forms are codominant, it can be seen that the chances of two unrelated
individuals having the same ‘set’ of HLA genes is very small, making the pos-sibility of finding ‘good’ transplant matches, other than identical twins, low.
In the HLA complex, the Class II region is located nearest to the centromere, which contains the DP, DQ and DR loci. Within each of these loci there are genes encoding the @ and A chains of the class II molecules. The situation is rather more complex than for Class I because each locus may contain more than one gene that encodes the @and A chains. For example, the HLA-DR region contains three or four genes for the HLA-A chain. All of the A gene products may be expressed in a single cell, making the degree of variation much higher than that of class I proteins. Like Class I, the Class II region also displays a high degree of polymorphism (Table 6.14).
HLA TYPING
Human Leukocyte Antigen typing is the process whereby the HLA antigens present on the cells of the recipient and the potential donor are determined. By determining the HLA types of potential transplant recipients, and storing these details on computer databases it is possible to match the donor organ, when one becomes available, to the most appropriate recipient. The degree of match required depends on the organ being transplanted, and is much more stringent for bone marrow transplants than for solid organ transplants. The transplantation of bone marrow is used to treat a number of condi-tions, including immunodeficiency and some kinds of cancer . Transplantation of bone marrow presents particular difficulties because bone marrow contains immunocompetent cells that can lead to fatal GVHD.
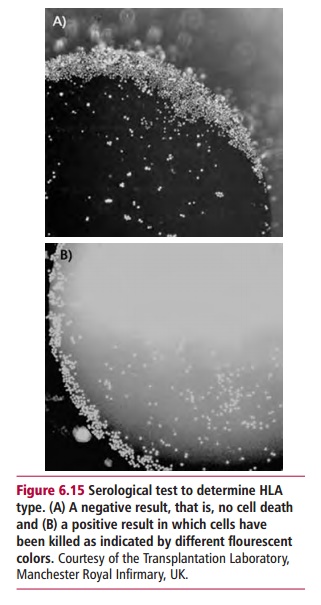
The traditional method for HLA typing is a serological technique that uses antibodies to known HLA antigens. This method is still used but is being superseded by molecular biological techniques. The serological method is the lymphocytotoxicity assay. In this assay, the cells to be typed are peripheral blood mononuclear cells (PBMC) that are readily obtained from whole blood. For typing Class II HLA antigens it is necessary to use purified B lymphocytes, since resting T lymphocytes do not express Class II molecules. Aliquots of the cells to be typed are pipetted into the wells of 96-well trays, known as Teresaki plates, after the inventor of this test. Antibodies to individual HLA antigens are added to each well and will bind to the lymphocytes if they express the appropriate antigen. The addition of complement to all the wells results in the death of cells with bound antibody. Viability stains, such as the fluores-cent acridine orange and ethidium bromide are then used to reveal wells containing dead cells. Acridine orange enters living cells and stains the nuclei green, while ethidium bromide enters dead cells and stains the nuclei red (Figure 6.15). The anti-HLA antibodies used in these tests may be obtained from people already sensitized to HLA antigens or they may be monoclonal antibodies with specificities for known HLA antigens. Rabbit serum is used as a source of complement. A test is scored as strongly positive when more than 50% of the cells in a well are killed.
Molecular biology techniques
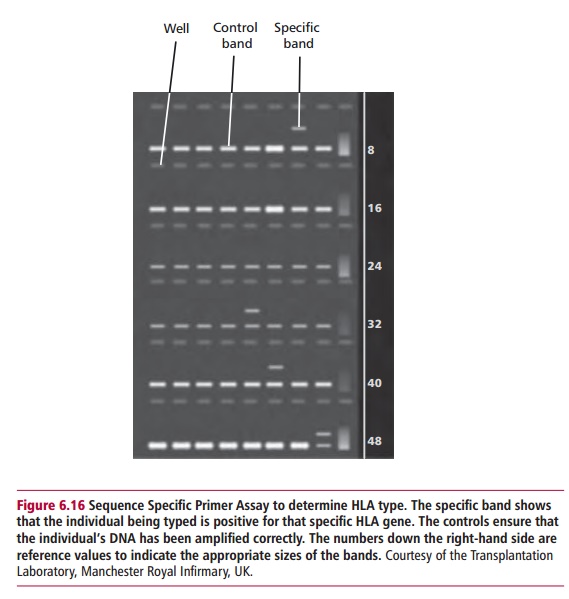
The main molecular biological technique used in histocompatibility testing is the polymerase chain reaction in which samples of DNA from the individual being typed is amplified many times over. Methods that use PCR require very little DNA and do not require living cells. For example, they can be carried out on whole blood that has been stored frozen. In the sequence specific primer (SSP) assay the primers used in the PCR are those specific for individual HLA alleles. The DNA will only be amplified in those mixtures that contain a probe complementary to that of the DNA being typed (Figure 6.16). Another method, called a sequence specific oligonucleotide probe (SSOP) assay, amplifies all the DNA by PCR and then uses sequence specific oligonucleotide probes to identify the products. In some laboratories the oligonucleotide probes are attached to microspheres. Incubation of the microspheres with the PCR product allows the latter to bind to any comple-mentary HLA sequence. The microspheres, with bound DNA, are then ana-lyzed in a flow cytometer to identify target HLA sequences.
The serological cross match
A serological cross match is carried out in order to detect the presence of anti-bodies to graft antigens in the serum of a potential recipient. Serum from the potential recipient is added to PBMC from the donor. Complement is added and the viability of the cells tested as previously. If donor cells are killed then it is probable that the recipient already has antibodies against graft antigens and the donor is unlikely to be a suitable match.
Matching donor and recipient
Individuals inherit a set of HLA genes from each parent. Thus, siblings are more likely to have a closer match than unrelated donors and, occasionally, people have donated a healthy kidney to a sibling with renal failure. Other sources of kidneys for transplantation include cadavers, often people who have been killed in accidents. The HLA types of potential recipients are stored in computer databases so that when a kidney donor becomes available, they can be HLA typed and the kidneys given to those recipients whose HLA anti-gens match as closely as possible.
Retrospective studies on the outcome of kidney transplantation have shown that matching the donor to the recipient improves graft survival. Thus, in the most favorable match, that is no mismatches of the HLA-DR, HLA-A and HLA-B alleles graft survival is superior to those with no mismatch of HLA-DR and only one mismatch of HLA-A and/or HLA-B alleles. These also gave significantly better graft survival than any other mismatched graft.
Related Topics