Chapter: Biotechnology Applying the Genetic Revolution: Transgenic Animals
Control by Site-Specific Recombination Using Cre or Flp
CONTROL
BY SITE-SPECIFIC RECOMBINATION USING CRE OR FLP
Another way to control the
expression of a transgene is via site-specific recombination. In this approach,
segments of DNA are physically removed or inverted to achieve activation of the
transgene. These DNA manipulations are done after the DNA carrying the
transgene and associated sequences has been successfully incorporated into the
germline chromosomes of the host animal.
Site-specific recombination
involves recognition of short specific sequences by DNA binding proteins.
Recombination then occurs between two of the recognition sequences. Some
site-specific recombination systems require several proteins for recognition and
crossing over. Others need only a single protein to bind the two recognition
sequences and recombine them. These are obviously far more useful in genetic
engineering.
Two such recombinase systems have been widely used: the Cre recombinase from bacterial virus P1 and the Flp recombinase (flippase) from the
2-micron plasmid of yeast. Both Cre and Flp recognize 34 base-pair sites (known
as loxP
and FRT, respectively) consisting of
13 base-pair inverted repeats flanking a central core of 8 base-pairs.
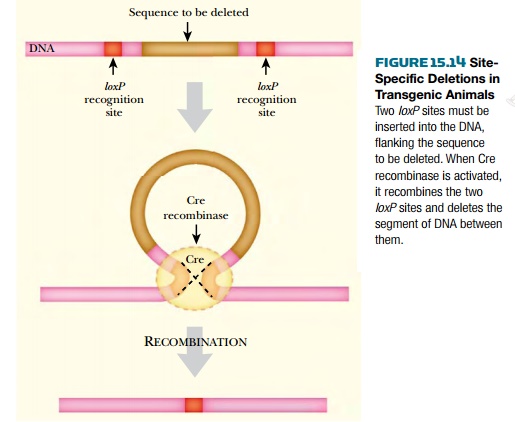
We have already described the
use of the Cre/loxP system in plants
to delete unwanted DNA segments after integration of incoming transgenic DNA.
Similar summarized as follows:
(a)
Removal of selective markers. Once transgenic DNA has been successfully
integrated, the antibiotic resistance gene used for selection and/or the reporter gene used for screening are no longer needed. If they are
enclosed between loxP or FRT sites,
they can be removed, leaving a transgenic organism with only the actual transgene
(plus a single copy of the loxP or
FRT site).
(b) Activation of transgene. Here the original transgenic construct is made with a blocking sequence between the
promoter and the transgene. The blocking sequence is flanked by loxP or FRT sites. After integration of
the incoming DNA, the blocking sequence is removed by Cre or Flp recombinase,
thus activating the transgene. deletions or rearrangements of eukaryotic
chromosomes may be generated in vivo
by using the Cre/loxP system. Two loxP sites are introduced by two rounds
of DNA insertion at separate specific locations. Then the Cre recombinase is
activated and deletions are generated.
Creation of conditional
knockout mutants. Transgenic constructs may be designed so that a specific gene
can be deleted in vivo. Generally,
two loxP sites are inserted into the
introns flanking an essential exon of the target gene. On recombination, the
exon will be deleted and the target gene will be inactivated. This allows
investigation of genes whose knockout mutations are lethal at the embryonic
When these recombinase systems are used, the recognition sequences are included
in the transgenic constructs. Later, the recombinase itself is provided by one
of three methods:stage. The animal can be allowed to grow into an adult before
the recombinase is activated to generate the knockout.
When these recombinase
systems are used, the recognition sequences are included in the transgenic
constructs. Later, the recombinase itself is provided by one of three methods:
(a)
The gene for recombinase may be carried on a plasmid that is
transformed into the animal. The recombinase will be expressed transiently,
assuming the plasmid does not integrate or survive over the long term.
(b)
The recombinase gene may itself be part of the transgenic construct
and be induced by some external stimulus.
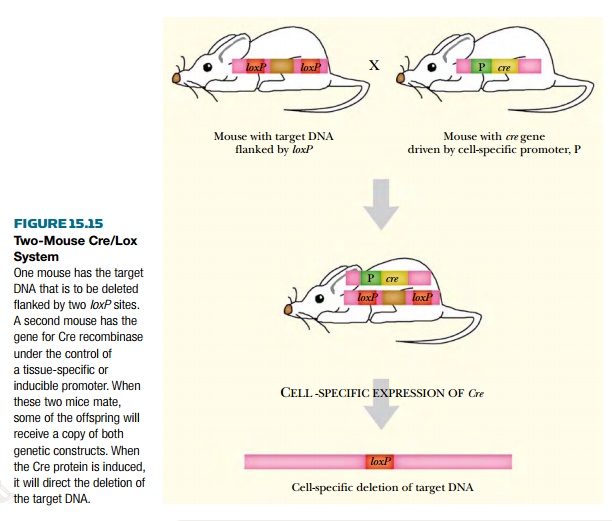
(c)
Two separate lines of transgenic organisms are used. The transgene
plus recognition sites are present in one host line and a second line of
transgenic organisms expresses the recombinase (Fig. 15.15). The two lines are
then mated together and the deletions occur in their progeny.
This approach can save a lot
of work. Instead of making separate transgenic constructs for each gene under
each condition, two sets of transgenic animals, usually mice, are generated and
are then crossed to investigate a wide range of genes and environmental
conditions. One set of mouse lines has Cre under the control of a variety of
promoters that are specific for different tissues or induced by different
signals. The second set of mouse lines has a series of different target genes
flanked by loxP sites. Thus the role
of any particular target gene may be investigated under any of the available
conditions by crossing the appropriate pair of strains.
Related Topics