Chapter: Biotechnology Applying the Genetic Revolution: Immune Technology
Identifying New Antigens for Vaccines
IDENTIFYING
NEW ANTIGENS FOR VACCINES
Another approach to creating
vaccines is to identify bacterial pathogen genes that are expressed when the
pathogen enters the host. These usually encode proteins that are different from
surface antigens. They encompass a variety of adaptations the pathogen makes in
order to live within the host organism.
Typically bacteria that enter
animals are engulfed by phagocytes and digested by the enzymes within the
lysosome. Some pathogens are engulfed as usual, but avoid digestion.
Modifications required to live intracellularly include changes in nutrition and
metabolism and mechanisms to protect against host attacks. Many different types
of genes are needed for this switch, and the products of these genes are
potential antigens for vaccine development.
Traditionally, identifying
genes that are expressed only in host cells relies on gene fusions. Suspected
genes are genetically fused to a reporter such as β-galactosidase, luciferase, or green
fluorescent protein (GFP), or to epitope tags such as FLAG or myc. The fusion
gene is introduced into the pathogenic organism, which is then allowed to
infect the host. The amount of reporter gene expression correlates with the
expression level of the target gene. For example, if the target gene is linked
to GFP, the amount of fluorescence is monitored with fluorescence microscopy or
fluorescence-based flow cytometry. If the fluorescence increases with host cell
invasion, then the target gene is a potential vaccine candidate because it may
be important for bacterial pathogenesis.
Individual gene fusions are
fine for suspected genes, but screening for novel genes with this method would
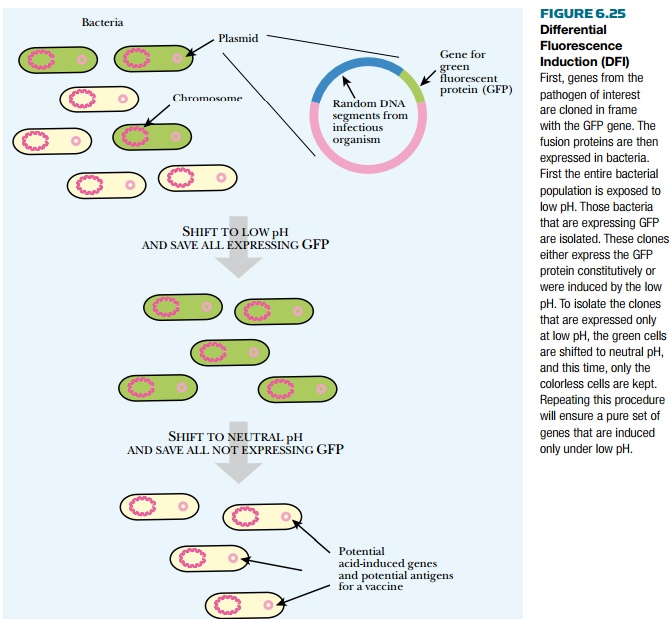
be tedious. Instead, differential fluorescence induction (DFI) uses a
combination of GFP fusions and FACS sorting (see earlier discussion) to
identify novel genes involved with host invasion (Fig. 6.25). First, a library
of genes from the pathogenic organism is genetically linked to GFP. The library
is transformed into bacterial cells where the gene fusions are expressed to
give GFP. The bacteria are then given a specific stimulus related to host
invasion. For example, when phagocytes engulf them, bacteria leave a neutral
environment (pH 7) and enter a compartment that is acidic (pH 4). To determine
if pH change induces gene expression, the bacteria with the fusion library are
shifted to an acidic environment. They are then sorted using FACS to collect
those with high GFP expression. If the novel gene fused to GFP is truly induced
by low pH, its GFP levels should drop when it is shifted back to neutral pH.
Therefore the cells with high GFP expression are shifted to pH 7 and resorted,
but this time bacteria with low levels of GFP are collected. The smaller pool
of bacteria are again stimulated with low pH and sorted, collecting those with
high GFP expression. This sorting scheme eliminates genes that are
constitutively expressed plus those that are not induced by low pH. The
remaining genes are acid-induced genes that adapt the organism to living within
the host. These may then be evaluated as antigens for vaccine development.
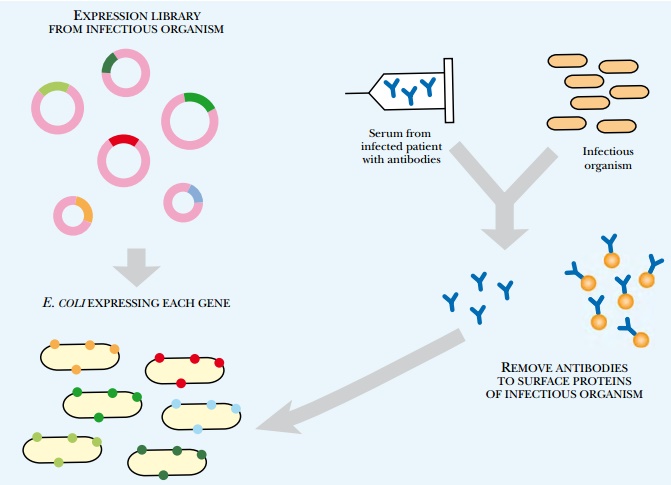
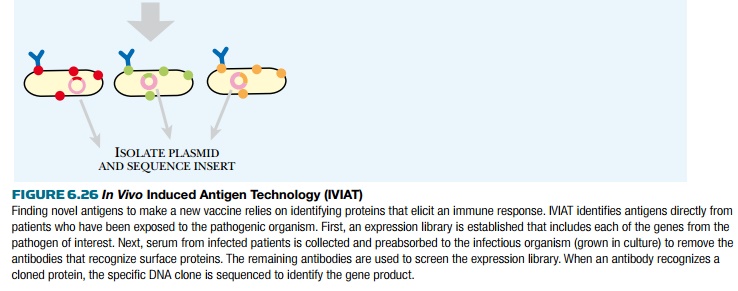
Another method to identify new antigens for vaccine development is in vivo induced antigen technology (IVIAT; Fig. 6.26). This method takes serum from patients who have been infected with a particular disease to which a vaccine is needed. The serum is a rich source of antibodies against the chosen disease agent. The serum is then mixed with a sample of the disease-causing microorganism. This removes antibodies that bind to proteins expressed by the microorganism while outside the host. This leaves a pool of antibodies against proteins that are expressed only during infection. To identify the proteins corresponding to these antibodies, a genomic expression library is constructed containing all the genes from the microorganism. The library is expressed in E. coli and is probed by the remaining antibodies. When an antibody matches a library clone, the gene insert is sequenced to identify the protein antigen. This method directly identifies protein antigens that stimulated antibody production during a genuine infection; therefore, antigens identified by this method are likely vaccine candidates.
Related Topics