Mechanism, Types, Importance, Recombination - Botany : Chromosomal Basis of Inheritance - Crossing Over | 12th Botany : Chapter 3 : Chromosomal Basis of Inheritance
Chapter: 12th Botany : Chapter 3 : Chromosomal Basis of Inheritance
Crossing Over
Crossing
Over
Crossing over is a
biological process that produces new combination of genes by inter-changing the
corresponding segments between non-sister chromatids of homologous pair of
chromosomes. The term 'crossing over' was coined by Morgan (1912). It
takes place during pachytene stage of prophase I of meiosis. Usually
crossing over occurs in germinal cells during gametogenesis. It is called
meiotic or germinal crossing over. It has universal occurrence and has great
significance. Rarely, crossing over occurs in somatic cells during mitosis. It
is called somatic or mitotic crossing over.
1. Mechanism of Crossing Over
Crossing over is a
precise process that includes stages like synapsis, tetrad formation, cross
over and terminalization.
(i) Synapsis
Intimate pairing between
two homologous chromosomes is initiated during zygotene stage of prophase I of
meiosis I. Homologous chromosomes are aligned side by side resulting in a pair
of homologous chromosomes called bivalents. This pairing phenomenon is
called synapsis or syndesis. It is of three types,
1. Procentric synapsis: Pairing starts from middle
of the chromosome.
2. Proterminal synapsis:
Pairing starts from the
telomeres.
3. Random synapsis: Pairing may start from
anywhere.
(ii) Tetrad Formation
Each homologous
chromosome of a bivalent begin to form two identical sister chromatids, which
remain held together by a centromere. At this stage each bivalent has four
chromatids. This stage is called tetrad stage.
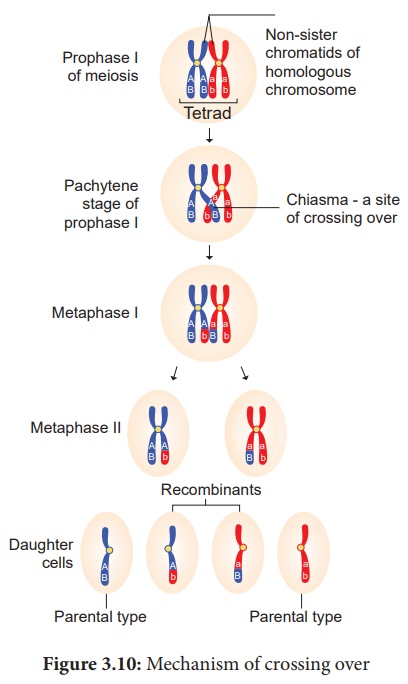
(iii) Cross Over
After tetrad formation,
crossing over occurs in pachytene stage. The non-sister chromatids of
homologous pair make a contact at one or more points. These points of contact
between non-sister chromatids of homologous chromosomes are called Chiasmata
(singular-Chiasma). At chiasma, cross-shaped or X-shaped structures are formed,
where breaking and rejoining of two chromatids occur. This results in
reciprocal exchange of equal and corresponding segments between them. A recent
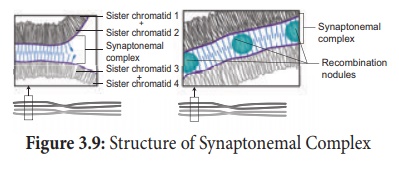
study reveals that synapsis and chiasma formation are facilitated by a highly
organised structure of filaments called Synaptonemal Complex (SC) (Figure
3.9). This synaptonemal complex formation is absent in some species of
male Drosophila hence crossing over does not takes place.
(iv) Terminalisation
After crossing over,
chiasma starts to move towards the terminal end of chromatids. This is known as
terminalisation. As a result, complete separation of homologous
chromosomes occurs. (Figure 4.10)
2. Types of Crossing Over
Depending upon the
number of chiasmata formed crossing over may be classified into three types.
(Figure 3.11)
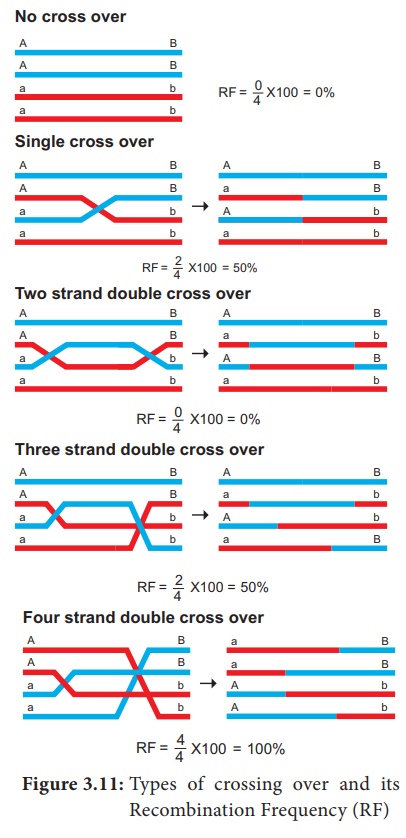
1. Single cross over: Formation of single
chiasma and involves only two chromatids out of four.
2. Double cross over: Formation of two chiasmata
and involves two or three or all four strands
3. Multiple cross over: Formation of more than
two chiasmata and crossing over frequency is extremely low.
3. Importance of Crossing Over
Crossing over occurs in
all organisms like bacteria, yeast, fungi, higher plants and animals. Its
importance is
·
Exchange of segments leads to new gene combinations which plays an
important role in evolution.
·
Studies of crossing over reveal that genes are arranged linearly
on the chromosomes.
·
Genetic maps are made based on the frequency of crossing over.
·
Crossing over helps to understand the nature and mechanism of gene
action.
·
If a useful new combination is formed it can be used in plant
breeding.
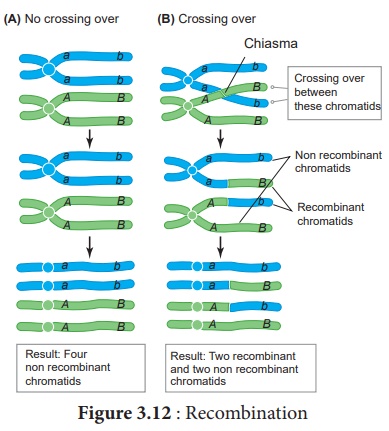
4. Recombination
Crossing over results in
the formation of new combination of characters in an organism called recombinants.
In this, segments of DNA are broken and recombined to produce new combinations
of alleles. This process is called Recombination. (Figure 3.12)
The widely accepted
model of DNA recombination during crossing over is Holliday’s hybrid DNA
model. It was first proposed by Robin Holliday in 1964. It
involves several steps. (Figure 3.13)
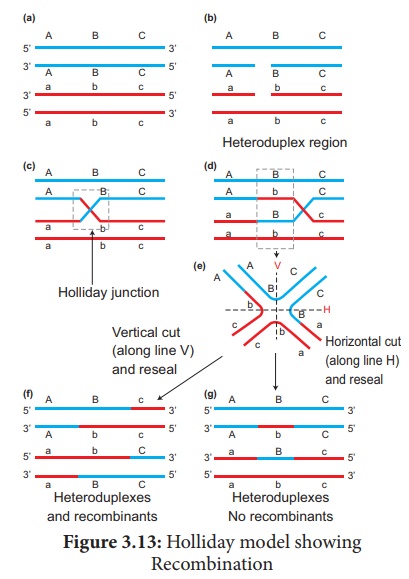
1. Homologous DNA molecules
are paired side by side with their duplicated copies of DNAs
2. One strand of both DNAs
cut in one place by the enzyme endonuclease.
3. The cut strands cross
and join the homologous strands forming the Holliday structure or
Holliday junction.
4. The Holliday junction
migrates away from the original site, a process called branch migration,
as a result heteroduplex region is formed.
5. DNA strands may cut
along through the vertical (V) line or horizontal (H) line.
6. The vertical cut will result
in hetero duplexes with recombinants.
7. The horizontal cut will
result in hetero duplex with non recombinants.
Calculation of
Recombination Frequency (RF)
The percentage of recombinant
progeny in a cross is called recombination frequency. The recombination
frequency (cross over frequency) (RF) is calculated by using the following
formula. The data is obtained from alleles in coupling configuration (Figure
3.14)
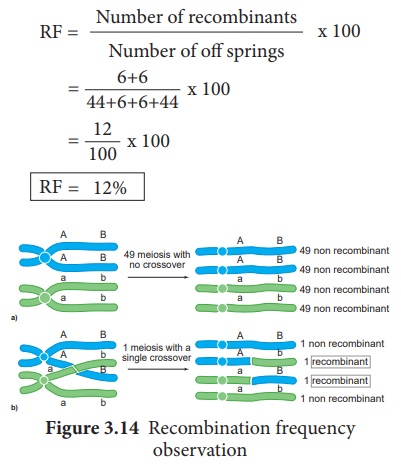
5. Genetic Mapping
Genes are present in a
linear order along the chromosome. They are present in a specific location
called locus (plural: loci). The diagrammatic representation of position
of genes and related distances between the adjacent genes is called genetic
mapping. It is directly proportional to the frequency of
recombination between them. It is also called as linkage map. The
concept of gene mapping was first developed by Morgan’s student Alfred H
Sturtevant in 1913. It provides clues about where the genes lies on
that chromosome.
Map distance
The unit of distance in
a genetic map is called a map unit (m.u). One map unit is equivalent to
one percent of crossing over (Figure 4. ). One map unit is also called a
centimorgan (cM) in honour of T.H. Morgan. 100 centimorgan is equal to
one Morgan (M). For example: A distance between A and B genes is estimated to
be 3.5 map units. It is equal to 3.5 centimorgans or 3.5 % or 0.035
recombination frequency between the genes.
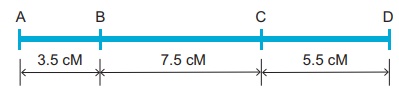
Genetic maps can be constructed
from a series of test crosses for pairs of genes called two point
crosses. But this is not efficient because double cross over is
missed.
Three point test cross
A more efficient mapping
technique is to construct based on the results of three-point test cross. It
refers to analyzing the inheritance patterns of three alleles by test crossing
a triple recessive heterozygote with a triple recessive homozygote. It enables
to determine the distance between the three alleles and the order in which they
are located on the chromosome. Double cross overs can be detected which will
provide more accurate map distances.
Three-point test cross can be best understood by considering following an example.
In maize (corn), the
three recessive alleles are
1. l for lazy or prostrate growth habit
2. g for glossy leaf
3. s for sugary endosperm
These three recessive
alleles (l g s) are crossed with wild type dominant alleles (L G S).
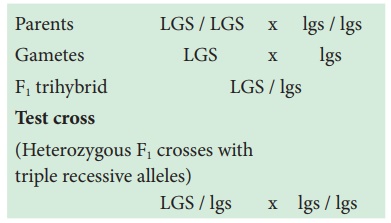
Parents LGS / LGS x lgs / lgs
Gametes LGS x lgs
F1 trihybrid LGS / lgs
Test cross
(Heterozygous F1
crosses with
triple recessive
alleles)
LGS / lgs x lgs / lgs
This trihybrid test
cross produces 8 different types (23=8) of gametes in which 740 progenies are
observed. The following table shows the result obtained from a test cross of
corn with three linked genes.
The analysis of a
three-point cross:
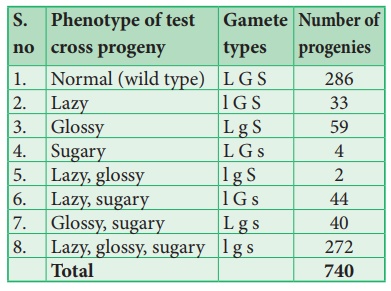
From the above result,
we must be careful to observe parental (P) and recombinant (R) types. First
note that parental genotypes for the triple homozygotes are L G S and l g s,
then analyse two recombinant loci at a time orderly L G/ l g, L S/ l s and G S/
g s. In this any combination other than these two constitutes a recombinant
(R).
Let’s analyse the loci
of two alleles at a time starting with L and G Since the L G and l g parental
genotypes the recombinants will be L g and l G. The Recombinant frequency (RF)
for these two alleles can be calculated as follows
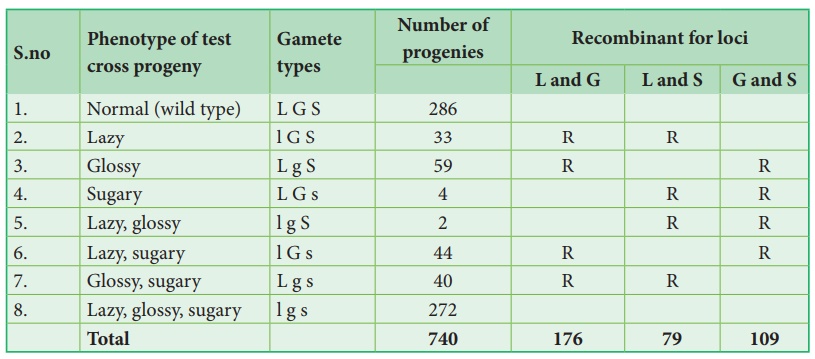
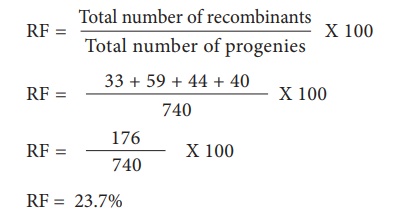
For L and S loci, the
recombinants are L s and l S. The Recombinant frequency (RF) will be as follows
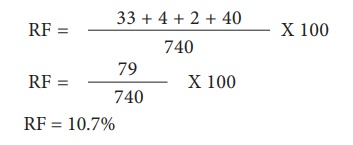
For G and S loci, the
recombinants are G s and g S. The Recombinant frequency (RF) will be as follows
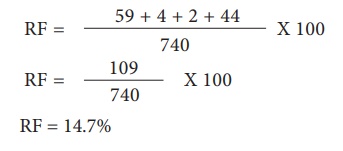
All the loci are linked,
because all the RF values are considerably less than 50%. In this L G loci show
highest RF value, they must be farthest apart. Therefore, the S locus must lie
between them. The order of genes should be l s g. A genetic map can be drawn as
follows: (Figure 3.15)
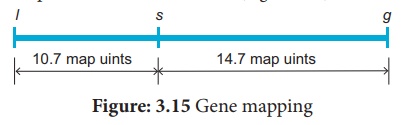
A final point note that
two smaller map distances, 10.7 m.u and 14.7., is add up to 25.4 m.u., which is
greater than 23.7 m.u., the distance calculated for l and g. we must identify
the two least number of progenies (totaling 8) in relation to recombination of
L and G. These two least progenies are double recombinants arising from double
cross over. The two least progenies not only counted once should have counted
each of them twice because each represents a double recombinant progeny.
Hence, we can correct
the value adding the numbers 33+59+44+40+4+4+2+2=188. Of the total of 740, this
number exactly 25.4 %, which is identical with the sum of two component values.
The test cross parental
combination can be re written as follows:
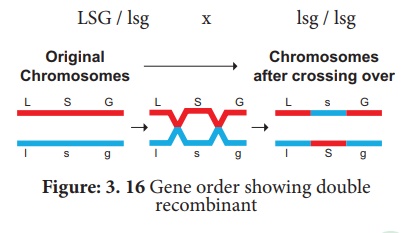
Uses of genetic mapping
·
It is used to determine gene order, identify the locus of a gene
and calculate the distances between genes.
·
They are useful in predicting results of dihybrid and trihybrid
crosses.
·
It allows the geneticists to understand the overall genetic
complexity of particular organism.
Related Topics